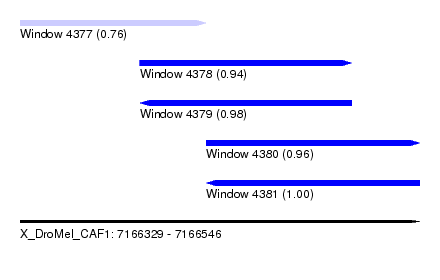
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,166,329 – 7,166,546 |
| Length | 217 |
| Max. P | 0.997713 |

| Location | 7,166,329 – 7,166,430 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.56 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7166329 101 + 22224390 AACAGCGAUGUGUGGCAUUUUCUAACAGUUGCUGCUGCU----GUCGCAGGUUGCAGGUUGCAUGUUUUGUUGCAGU----UGCUGCAGCUUAUAUGACGCUUACAUUU ....(((((.(((((((........((((....)))).)----)))))).)))))((((..(((((...(((((((.----..)))))))..)))))..))))...... ( -32.80) >DroSec_CAF1 33971 94 + 1 AACAGCGAUGUGUGGCAUUUUCCAACAGUUGCUGCUGUU----GUCGCAGGUUGCCGG-------UUUUGUGGCAGU----UGCUGCAGUUUCUAUGACGCUUGCAUUU ....((((.((((((((.....(((((((....))))))----)........))))((-------..(((..((...----.))..)))..))....)))))))).... ( -30.82) >DroSim_CAF1 28184 94 + 1 AACAGCGAUGUGUGGCAUUUUCUAAUAGUUGCUGCUGCU----GUCGCAGGUUGCAGG-------UUUUGUGGCAGU----UGCUGCAGUUUCUAUGACGCUUACAUUU ...(((((((.....)))......((((..(((((.(((----((((((((.......-------.)))))))))))----....)))))..))))..))))....... ( -30.10) >DroEre_CAF1 34824 85 + 1 AACAGCGAUGUGUGGCAUUUUCCAAGAGUUGCUG----------UCGCAGGUUGCAGG-------CUUUGUGGCAGU-------UGCAGGUUCUAUGACGCUUGCAUUU .((((((((...(((......)))...)))))))----------).((((((((((.(-------((....)))...-------)))).(((....))))))))).... ( -27.20) >DroYak_CAF1 35960 99 + 1 AACAGCGAUGUGUGGCAUUUUCUAACAGUUGCUGCAGUCGCAGGUUGCAGGUUGCAGG-------CUUUGUGGCAGUUCUAUGCUGCAGGUUCUGUGACGCUUG---UU (((((((((.((..(((............)))..))))))).....((..((..(((.-------.((((..(((......)))..))))..)))..)))).))---)) ( -36.10) >consensus AACAGCGAUGUGUGGCAUUUUCUAACAGUUGCUGCUGCU____GUCGCAGGUUGCAGG_______UUUUGUGGCAGU____UGCUGCAGGUUCUAUGACGCUUGCAUUU ..(((((((...(((......)))...)))))))............((((((.(((((........))))).(((((.....)))))............)))))).... (-19.60 = -20.56 + 0.96)

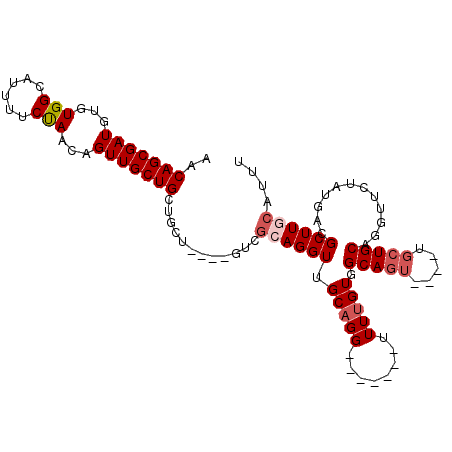

| Location | 7,166,394 – 7,166,509 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.45 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -14.67 |
| Energy contribution | -15.85 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7166394 115 + 22224390 GUUGCAGU----UGCUGCAGCUUAUAUGACGCUUACAUUUGUUAUAUGUAUCUGCCACUGGGGGUUUUCCGUCAAUAUUGCAGGUUGCAUAGCUUAUGCUGCAGCUUCUAUU-GAUGACU ((((((((----.((((((((..((((((((........))))))))((((((((...(((.((....)).))).....))))).))).........))))))))....)))-).)))). ( -35.90) >DroSec_CAF1 34029 115 + 1 GUGGCAGU----UGCUGCAGUUUCUAUGACGCUUGCAUUUGUUAUAUGUAUCUGCCACUAGGGGUUUUCCGUCGAUGUUGCAGGUUGCAUAGCUAAUGCUGCAACUUCUAUU-GAUAACU ((((.(((----(((.((.(((.....)))))..(((((.((..(((((((((((.((..((((....)).))...)).))))).)))))))).))))).)))))).)))).-....... ( -32.80) >DroSim_CAF1 28242 115 + 1 GUGGCAGU----UGCUGCAGUUUCUAUGACGCUUACAUUUGUUAUAAGUAUCUGCCACUAGGGGUUUUCCAUCGAUGUUGCAGGUUGCAUAGCUUAUGCUGCAACUUCUAUU-GAUAACU (((((((.----((((........(((((((........))))))))))).)))))))..(((....)))(((((((..(.((((((((..((....)))))))))))))))-))).... ( -33.30) >DroEre_CAF1 34876 95 + 1 GUGGCAGU-------UGCAGGUUCUAUGACGCUUGCAUUUGGU----GUAUCUGCUGCUGGGA-UUUUUCCUGGAUGUCGCAGGUUGCAUAGCUUAUGCUGCAGCUA------------- ((((((..-------.((((((......(((((.......)))----))))))))..(..(((-....)))..).)))))).(((((((..((....))))))))).------------- ( -33.60) >DroYak_CAF1 36022 101 + 1 GUGGCAGUUCUAUGCUGCAGGUUCUGUGACGCUUG---UUGCU----GUG-----------CG-UUGAUGUUGCAGGUUGCAGGUUGCACAGCUUGUGUGGCAGCUUCCCUAGGAUGACG (..(((......)))..)(((..((((.((((...---..(((----(((-----------((-....(((........)))...))))))))..)))).))))....)))......... ( -35.30) >consensus GUGGCAGU____UGCUGCAGGUUCUAUGACGCUUGCAUUUGUUAUA_GUAUCUGCCACUAGGGGUUUUCCGUCGAUGUUGCAGGUUGCAUAGCUUAUGCUGCAGCUUCUAUU_GAUAACU ...(((((.....)))))............((..(((((...........(((......)))...........))))).))(((((((..(((....))))))))))............. (-14.67 = -15.85 + 1.18)

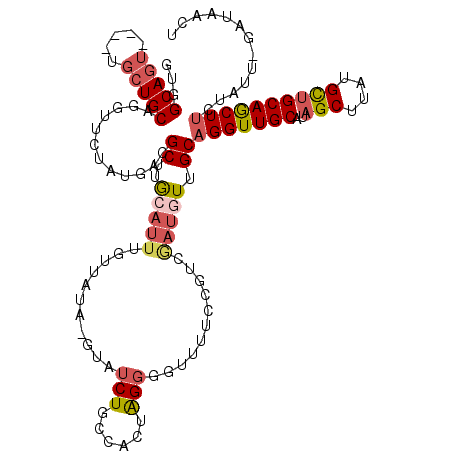

| Location | 7,166,394 – 7,166,509 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.45 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.76 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7166394 115 - 22224390 AGUCAUC-AAUAGAAGCUGCAGCAUAAGCUAUGCAACCUGCAAUAUUGACGGAAAACCCCCAGUGGCAGAUACAUAUAACAAAUGUAAGCGUCAUAUAAGCUGCAGCA----ACUGCAAC .......-..(((..((((((((.((....((((...((((..(((((..((.....)).))))))))).(((((.......))))).))))....)).)))))))).----.))).... ( -31.20) >DroSec_CAF1 34029 115 - 1 AGUUAUC-AAUAGAAGUUGCAGCAUUAGCUAUGCAACCUGCAACAUCGACGGAAAACCCCUAGUGGCAGAUACAUAUAACAAAUGCAAGCGUCAUAGAAACUGCAGCA----ACUGCCAC .(((.((-....((.((((((((((.....))))....)))))).))....)).))).....(((((((...(((.......)))...((..((.......))..)).----.))))))) ( -26.10) >DroSim_CAF1 28242 115 - 1 AGUUAUC-AAUAGAAGUUGCAGCAUAAGCUAUGCAACCUGCAACAUCGAUGGAAAACCCCUAGUGGCAGAUACUUAUAACAAAUGUAAGCGUCAUAGAAACUGCAGCA----ACUGCCAC .(((.((-....((.(((((((((((...)))))....)))))).))....)).))).....(((((((...((((((.....)))))).((..(((...)))..)).----.))))))) ( -28.80) >DroEre_CAF1 34876 95 - 1 -------------UAGCUGCAGCAUAAGCUAUGCAACCUGCGACAUCCAGGAAAAA-UCCCAGCAGCAGAUAC----ACCAAAUGCAAGCGUCAUAGAACCUGCA-------ACUGCCAC -------------..(((((.(((((...)))))..((((.(....))))).....-.....)))))......----......((((.(.((......)))))))-------........ ( -21.90) >DroYak_CAF1 36022 101 - 1 CGUCAUCCUAGGGAAGCUGCCACACAAGCUGUGCAACCUGCAACCUGCAACAUCAA-CG-----------CAC----AGCAA---CAAGCGUCACAGAACCUGCAGCAUAGAACUGCCAC ..((.(((...))).(((((.......(((((((....(((.....))).......-.)-----------)))----)))..---..((.((......)))))))))...))........ ( -22.50) >consensus AGUCAUC_AAUAGAAGCUGCAGCAUAAGCUAUGCAACCUGCAACAUCGACGGAAAACCCCCAGUGGCAGAUAC_UAUAACAAAUGCAAGCGUCAUAGAAACUGCAGCA____ACUGCCAC ...............(((((((......(((((.....(((.........((.....))......(((...............)))..))).)))))...)))))))............. (-11.46 = -11.76 + 0.30)

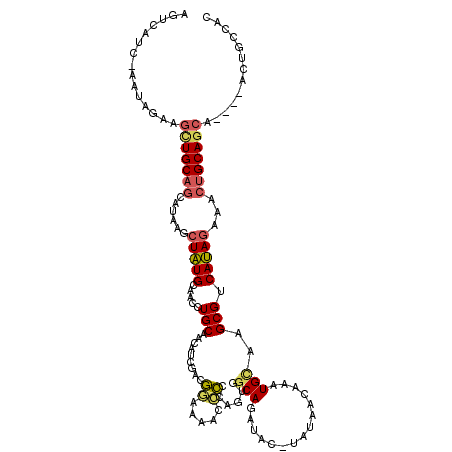

| Location | 7,166,430 – 7,166,546 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.76 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7166430 116 + 22224390 GUUAUAUGUAUCUGCCACUGGGGGUUUUCCGUCAAUAUUGCAGGUUGCAUAGCUUAUGCUGCAGCUUCUAUU-GAUGACUAAUU---UAUUGUUGCUUGCUUGCUUGAGCAAGUUGCUUU (((((......((((...(((.((....)).))).....))))((((((..((....)))))))).......-.))))).....---....((.((((((((....)))))))).))... ( -31.50) >DroSec_CAF1 34065 116 + 1 GUUAUAUGUAUCUGCCACUAGGGGUUUUCCGUCGAUGUUGCAGGUUGCAUAGCUAAUGCUGCAACUUCUAUU-GAUAACUAAUU---UGUUGUUGCUUGAUUGCUUAAGCAAGUGGCUUU .............((((((...((((.((.((.((.((((((((((((...)).))).))))))).)).)).-)).))))....---......(((((((....)))))))))))))... ( -32.20) >DroSim_CAF1 28278 116 + 1 GUUAUAAGUAUCUGCCACUAGGGGUUUUCCAUCGAUGUUGCAGGUUGCAUAGCUUAUGCUGCAACUUCUAUU-GAUAACUAAUU---UGUUGUUGCUUGAUUGCUUAAGCAAGUGGCUUU .............((((((((.((....))(((((((..(.((((((((..((....)))))))))))))))-)))..)))...---.....((((((((....)))))))))))))... ( -31.80) >DroEre_CAF1 34909 95 + 1 GGU----GUAUCUGCUGCUGGGA-UUUUUCCUGGAUGUCGCAGGUUGCAUAGCUUAUGCUGCAGCUA-------------------AUAUUGUUGCAUGAAUGUGUCAGCAAGUGGCU-C .((----((((((((..(..(((-....)))..).....))))).)))))(((....)))..(((((-------------------...(((((((((....))).)))))).)))))-. ( -32.40) >DroYak_CAF1 36059 104 + 1 GCU----GUG-----------CG-UUGAUGUUGCAGGUUGCAGGUUGCACAGCUUGUGUGGCAGCUUCCCUAGGAUGACGCAUUCGCUGCUGCUGCUUGAAUGUUUCAAGAAGUGGCUUU ((.----(((-----------((-((...(((((....((((((((....))))))))..))))).(((...))).)))))))..)).((..((.(((((.....))))).))..))... ( -36.80) >consensus GUUAUA_GUAUCUGCCACUAGGGGUUUUCCGUCGAUGUUGCAGGUUGCAUAGCUUAUGCUGCAGCUUCUAUU_GAUAACUAAUU___UGUUGUUGCUUGAUUGCUUAAGCAAGUGGCUUU ...........((((.((...((.....))......)).))))(((((..(((....))))))))..........................((..((((..........))))..))... (-14.80 = -15.40 + 0.60)



| Location | 7,166,430 – 7,166,546 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.76 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -7.95 |
| Energy contribution | -7.95 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7166430 116 - 22224390 AAAGCAACUUGCUCAAGCAAGCAAGCAACAAUA---AAUUAGUCAUC-AAUAGAAGCUGCAGCAUAAGCUAUGCAACCUGCAAUAUUGACGGAAAACCCCCAGUGGCAGAUACAUAUAAC ...((..(((((....)))))...)).......---.....((((((-((((..((((........)))).(((.....))).)))))).((.......))..))))............. ( -22.50) >DroSec_CAF1 34065 116 - 1 AAAGCCACUUGCUUAAGCAAUCAAGCAACAACA---AAUUAGUUAUC-AAUAGAAGUUGCAGCAUUAGCUAUGCAACCUGCAACAUCGACGGAAAACCCCUAGUGGCAGAUACAUAUAAC ...(((((((((((.(....).)))))......---.....(((.((-....((.((((((((((.....))))....)))))).))....)).)))....))))))............. ( -28.90) >DroSim_CAF1 28278 116 - 1 AAAGCCACUUGCUUAAGCAAUCAAGCAACAACA---AAUUAGUUAUC-AAUAGAAGUUGCAGCAUAAGCUAUGCAACCUGCAACAUCGAUGGAAAACCCCUAGUGGCAGAUACUUAUAAC ...(((((((((((.(....).)))))......---.....(((.((-....((.(((((((((((...)))))....)))))).))....)).)))....))))))............. ( -28.80) >DroEre_CAF1 34909 95 - 1 G-AGCCACUUGCUGACACAUUCAUGCAACAAUAU-------------------UAGCUGCAGCAUAAGCUAUGCAACCUGCGACAUCCAGGAAAAA-UCCCAGCAGCAGAUAC----ACC .-......(((((((.....))).))))...(((-------------------(.(((((.(((((...)))))..((((.(....))))).....-.....))))).)))).----... ( -23.80) >DroYak_CAF1 36059 104 - 1 AAAGCCACUUCUUGAAACAUUCAAGCAGCAGCAGCGAAUGCGUCAUCCUAGGGAAGCUGCCACACAAGCUGUGCAACCUGCAACCUGCAACAUCAA-CG-----------CAC----AGC ..........(((((.....)))))..(((((.((....))....(((...))).))))).......(((((((....(((.....))).......-.)-----------)))----))) ( -25.20) >consensus AAAGCCACUUGCUGAAGCAAUCAAGCAACAACA___AAUUAGUCAUC_AAUAGAAGCUGCAGCAUAAGCUAUGCAACCUGCAACAUCGACGGAAAACCCCCAGUGGCAGAUAC_UAUAAC ........((((............(((((..........................))))).((((.....)))).....))))..................................... ( -7.95 = -7.95 + 0.00)

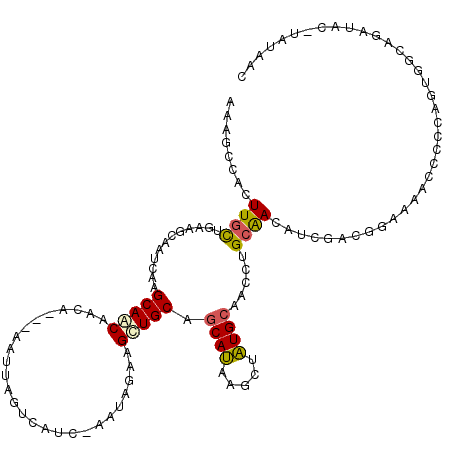

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:41 2006