
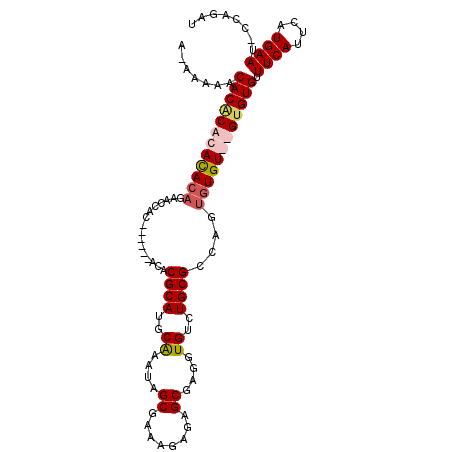
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,151,362 – 7,151,463 |
| Length | 101 |
| Max. P | 0.989787 |

| Location | 7,151,362 – 7,151,463 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -21.01 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7151362 101 + 22224390 AAAAAAUCACGCACACACAGAACCAC------ACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGGCAGUGUGUGUGUGUGUUUCAUUCAUGAAU-CCAGAU .....(((..(((((((((....(((------((.((((.(((....((........))....))).))))...))))))))))))))((((....)))).-...))) ( -31.30) >DroSec_CAF1 19098 100 + 1 A-AAAAUCACGCACACACAGAACCAC------ACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGGCAGUGUGUGUGUGUGUUUCAUUCAUGAAU-CCAGAU .-...(((..(((((((((....(((------((.((((.(((....((........))....))).))))...))))))))))))))((((....)))).-...))) ( -31.30) >DroEre_CAF1 19789 87 + 1 U-AAACACACACACACA------------------CGCAUGCGAAUAGCGAAAGAGAGCGAGGUGUUUGCGCCAGUGUGU--GCGUGUUUCAUUCAUGAAUUCCAGAU .-((((((.(((((((.------------------((((.(((....((........))....))).))))...))))))--).)))))).................. ( -28.00) >DroYak_CAF1 19419 105 + 1 A-AAAAACACACACAUACACACCCACAGUCAAACUCGCAUGCGAAUAGCAAAAGAGAGCGAGGUGUGUGCGCCAGUGUGU--GUGUGUUUCAUUCAUGAAUUCCAGAU .-..((((((((((((((...............(((...(((.....)))...))).....((((....)))).))))))--)))))))).................. ( -33.10) >consensus A_AAAAACACACACACACAGAACCAC______ACACGCAUGCAAAUAGCGAAAGAGAGCGAGGUGUCUGCGCCAGUGUGU__GUGUGUUUCAUUCAUGAAU_CCAGAU .......((((((((((((................((((..((....((........))....))..))))....)))))))))))).((((....))))........ (-21.01 = -22.20 + 1.19)


| Location | 7,151,362 – 7,151,463 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -14.90 |
| Energy contribution | -19.59 |
| Covariance contribution | 4.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7151362 101 - 22224390 AUCUGG-AUUCAUGAAUGAAACACACACACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUUGCAUGCGUGU------GUGGUUCUGUGUGUGCGUGAUUUUUU ......-.((((....)))).(((.((((((((..((((((.((.....((........)).....(....))).)------)))))..)))))))).)))....... ( -31.40) >DroSec_CAF1 19098 100 - 1 AUCUGG-AUUCAUGAAUGAAACACACACACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUUGCAUGCGUGU------GUGGUUCUGUGUGUGCGUGAUUUU-U ......-.((((....)))).(((.((((((((..((((((.((.....((........)).....(....))).)------)))))..)))))))).))).....-. ( -31.40) >DroEre_CAF1 19789 87 - 1 AUCUGGAAUUCAUGAAUGAAACACGC--ACACACUGGCGCAAACACCUCGCUCUCUUUCGCUAUUCGCAUGCG------------------UGUGUGUGUGUGUUU-A ..................((((((((--((((((..(((((........((........))........))))------------------)))))))))))))))-. ( -28.29) >DroYak_CAF1 19419 105 - 1 AUCUGGAAUUCAUGAAUGAAACACAC--ACACACUGGCGCACACACCUCGCUCUCUUUUGCUAUUCGCAUGCGAGUUUGACUGUGGGUGUGUAUGUGUGUGUUUUU-U .................(((((((((--(((.....((((((.(((.(((..(((...(((.....)))...)))..)))..))).)))))).)))))))))))).-. ( -34.20) >consensus AUCUGG_AUUCAUGAAUGAAACACAC__ACACACUGCCGCAGACACCUCGCUCUCUUUCGCUAUUCGCAUGCGUGU______GUGGUUCUGUGUGUGCGUGAUUUU_U .................((((((((((((((((..(((((........(((...................))).........)))))..))))))))))))))))... (-14.90 = -19.59 + 4.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:28 2006