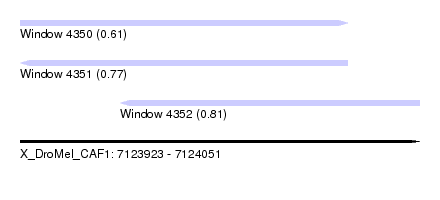
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,123,923 – 7,124,051 |
| Length | 128 |
| Max. P | 0.805563 |

| Location | 7,123,923 – 7,124,028 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.78 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
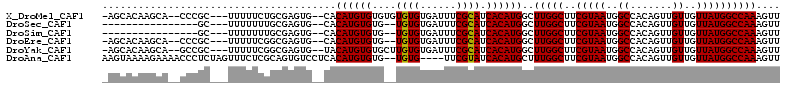
>X_DroMel_CAF1 7123923 105 + 22224390 -AGCACAAGCA--CCCGC---UUUUUCUGCGAGUG--CACAUGUGUGUGUGUGUGAUUUCGCAUCACAUGGCUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU -.((....(((--(.(((---.......))).)))--).((((((.(((((........)))))))))))))(((((..(((((..((.......))..)))))))))).... ( -37.70) >DroSec_CAF1 115000 90 + 1 ----------------GC---UUUUUUUGCGAGUG--CACAUGUGUG--UGUGUGAUUUCGCAUCACAUGGCUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU ----------------((---((...(((((((.(--(.((((((.(--((((......))))))))))).....)))))))))((((((.((......)).)))))))))). ( -29.90) >DroSim_CAF1 114637 90 + 1 ----------------GC---UUUUUUUGCGAGUG--CACAUGUGUG--UGUGUGAUUUCGCAUCACAUGGCUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU ----------------((---((...(((((((.(--(.((((((.(--((((......))))))))))).....)))))))))((((((.((......)).)))))))))). ( -29.90) >DroEre_CAF1 118222 103 + 1 -AGCACAAGCA--CCCGC---UUUUUCGGCGAGUG--CACAUGUGUG--UGUGUGAUUUCGCAUCACAUGGCUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU -.((....(((--(.(((---(.....)))).)))--).((((((.(--((((......)))))))))))))(((((..(((((..((.......))..)))))))))).... ( -38.40) >DroYak_CAF1 120360 105 + 1 -AGCACAAGCA--GCCGC---UUUUUCGGCGAGUG--UACAUGUGUGCUUGUGUGAUUUCGCAUCACAUGGCUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU -(((.(((((.--((((.---.....)))).....--..((((((....((((......)))).))))))))))))))......((((((.((......)).))))))..... ( -35.90) >DroAna_CAF1 166269 107 + 1 AAGUAAAAGAAAACCCUCUAGUUUCUCGCAGUGUCCUCACAUGUGUG--UGUG----UUCGUAUCACAUGCUUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU .......(((((.(......))))))(((((((....))).)))).(--((((----.......)))))((((((((..(((((..((.......))..))))))))))))). ( -26.30) >consensus _AGCACAAGCA__CCCGC___UUUUUCGGCGAGUG__CACAUGUGUG__UGUGUGAUUUCGCAUCACAUGGCUUGGCUUCGUAAUGGCCACAGUUGUUGUUAUGGCCAAAGUU .......................................((((((....((((......)))).))))))..(((((..(((((..((.......))..)))))))))).... (-21.92 = -21.78 + -0.14)


| Location | 7,123,923 – 7,124,028 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
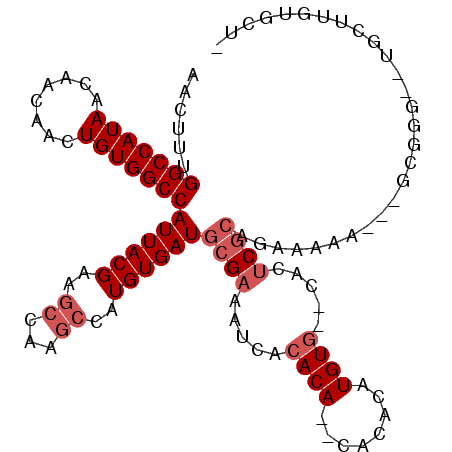
>X_DroMel_CAF1 7123923 105 - 22224390 AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACACACACACAUGUG--CACUCGCAGAAAAA---GCGGG--UGCUUGUGCU- .....((((((((........))))))))......((((((((..((((((.....))))))((((......))))--.((((((.......---)))))--)))))).)))- ( -36.60) >DroSec_CAF1 115000 90 - 1 AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA--CACACAUGUG--CACUCGCAAAAAAA---GC---------------- .....((((((((........)))))))).......((...((((((((.((.(........).--)))))))).)--)....)).......---..---------------- ( -23.30) >DroSim_CAF1 114637 90 - 1 AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA--CACACAUGUG--CACUCGCAAAAAAA---GC---------------- .....((((((((........)))))))).......((...((((((((.((.(........).--)))))))).)--)....)).......---..---------------- ( -23.30) >DroEre_CAF1 118222 103 - 1 AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA--CACACAUGUG--CACUCGCCGAAAAA---GCGGG--UGCUUGUGCU- .....((((((((........))))))))......((((((((..((((((.....))))))((--((....))))--.((((((.......---)))))--)))))).)))- ( -35.90) >DroYak_CAF1 120360 105 - 1 AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACAAGCACACAUGUA--CACUCGCCGAAAAA---GCGGC--UGCUUGUGCU- .....((((((((........))))))))......((((((((((((((.(((............)))))))))..--.....((((.....---.))))--.))))).)))- ( -37.70) >DroAna_CAF1 166269 107 - 1 AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAAGCAUGUGAUACGAA----CACA--CACACAUGUGAGGACACUGCGAGAAACUAGAGGGUUUUCUUUUACUU .....((((((((........))))))))...((...((...(((((((.......----....--..)))))))..))......)).((((((....))))))......... ( -24.22) >consensus AACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA__CACACAUGUG__CACUCGCAGAAAAA___GCGGG__UGCUUGUGCU_ ......(((((((........)))))))((((((..((...))..))))))((((.....((((........)))).....))))............................ (-17.50 = -18.50 + 1.00)


| Location | 7,123,955 – 7,124,051 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -14.08 |
| Energy contribution | -15.75 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
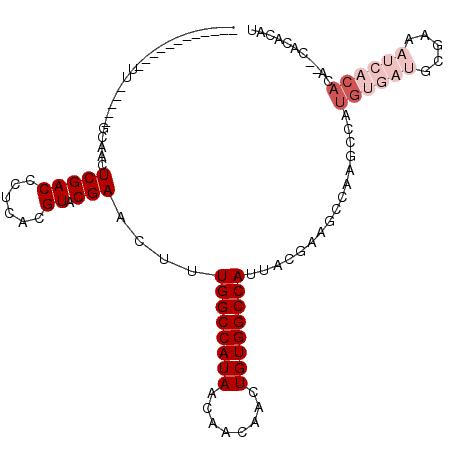
>X_DroMel_CAF1 7123955 96 - 22224390 ------------UU-----GCAACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACACACACACAU ------------..-----((...(((((......)).)))....((((((((........)))))))).......)).......((((((.....))))))........... ( -20.80) >DroSim_CAF1 114656 94 - 1 ------------UU-----GCAACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA--CACACAU ------------..-----((...(((((......)).)))....((((((((........)))))))).......)).......((((((.....))))))..--....... ( -20.80) >DroEre_CAF1 118254 94 - 1 ------------UU-----GCAACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA--CACACAU ------------..-----((...(((((......)).)))....((((((((........)))))))).......)).......((((((.....))))))..--....... ( -20.80) >DroWil_CAF1 226673 75 - 1 -------------G-----UCGACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAC------------------AUUCACACA--CACACAC -------------(-----(((...))))......((.((.....((((((((........))))))))...)).))------------------.........--....... ( -15.80) >DroYak_CAF1 120392 96 - 1 ------------UU-----GCAACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACAAGCACACAU ------------..-----((...(((((......)).)))....((((((((........)))))))).......))...((..((((((.....))))))...))...... ( -22.80) >DroAna_CAF1 166309 107 - 1 CUUCUUCGCUUUCUGCGACUCAACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAAGCAUGUGAUACGAA----CACA--CACACAU .....((((.....))))......(((....((((((.((.....((((((((........))))))))...))..((....))))))))..))).----....--....... ( -24.60) >consensus ____________UU_____GCAACUCGACCCUCACGUACGAACUUUGGCCAUAACAACAACUGUGGCCAUUACGAAGCCAAGCCAUGUGAUGCGAAAUCACACA__CACACAU ........................(((((......)).)))....((((((((........))))))))................((((((.....))))))........... (-14.08 = -15.75 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:15 2006