
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,109,715 – 7,109,821 |
| Length | 106 |
| Max. P | 0.988810 |

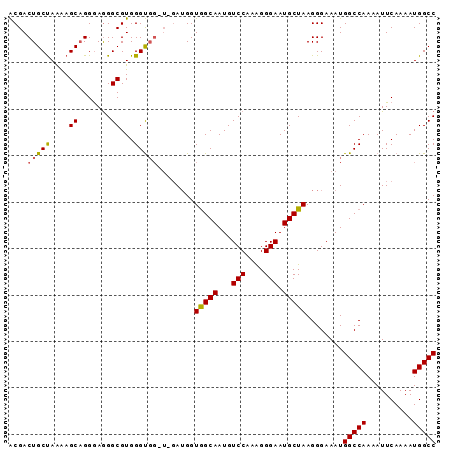
| Location | 7,109,715 – 7,109,807 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
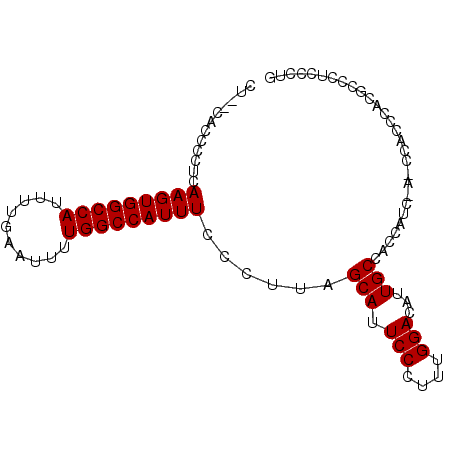
>X_DroMel_CAF1 7109715 92 + 22224390 ACGACUGCUAAAAGCUGGGAGGGCGUGGGUGGAUGGAUGGUAGCAAUGUCCAAAGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCC .((.(..((....(((.....)))...))..).)).....(((((...(((....))).)))))........(((((..........))))) ( -21.40) >DroSec_CAF1 100946 88 + 1 ACGACUGCUAAAAGCAGGGAGGGCGUGGGUGG----AUGGUGGCAAUGUCCAAAGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCC ....((((.....))))...((.(((...(((----((..((((....(((....((....))...)))....))))..)))))..))).)) ( -28.20) >DroEre_CAF1 103256 89 + 1 GCGACUGCCAGGAGCAGGAAAGGCGUGGGC---UUGAAGGUGGCAAUGUCCAAGGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCC (((.((((.....))))......))).(((---(((((..((((....(((...((.....))...)))....))))...))))....)))) ( -24.80) >consensus ACGACUGCUAAAAGCAGGGAGGGCGUGGGUGG_U_GAUGGUGGCAAUGUCCAAAGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCC ....(((((....((.......))...)))))........(((((...(((....))).)))))........(((((..........))))) (-18.69 = -18.70 + 0.01)


| Location | 7,109,715 – 7,109,807 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.27 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7109715 92 - 22224390 GGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCUUUGGACAUUGCUACCAUCCAUCCACCCACGCCCUCCCAGCUUUUAGCAGUCGU (((((..........)))))........(((((.(((....)))...))))).........................((.....))...... ( -16.40) >DroSec_CAF1 100946 88 - 1 GGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCUUUGGACAUUGCCACCAU----CCACCCACGCCCUCCCUGCUUUUAGCAGUCGU (((((..........)))))..........(((.(((....)))...)))......----...............((((.....)))).... ( -17.00) >DroEre_CAF1 103256 89 - 1 GGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCCUUGGACAUUGCCACCUUCAA---GCCCACGCCUUUCCUGCUCCUGGCAGUCGC (((...((((((...((((.((.(((...((.......)).))).)).))))..)))))---).....)))....((((.....)))).... ( -18.80) >consensus GGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCUUUGGACAUUGCCACCAUC_A_CCACCCACGCCCUCCCUGCUUUUAGCAGUCGU (((((..........)))))..........(((.(((....)))...))).........................((((.....)))).... (-15.43 = -15.77 + 0.33)

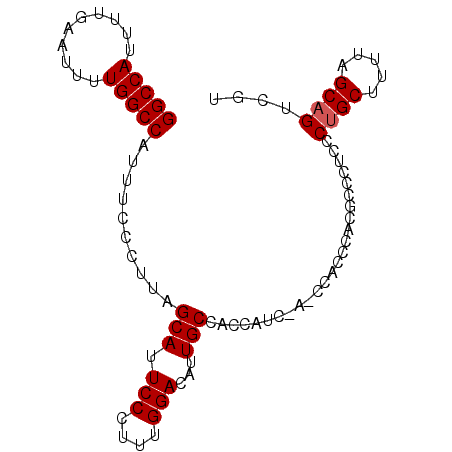

| Location | 7,109,729 – 7,109,821 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7109729 92 + 22224390 CUGGGAGGGCGUGGGUGGAUGGAUGGUAGCAAUGUCCAAAGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCCACUUGAGGGGUG--AG ((..((((((.....(((((...((((((((...(((....))).))))..........))))..))))).....))).)))..))....--.. ( -21.30) >DroSec_CAF1 100960 90 + 1 CAGGGAGGGCGUGGGUGG----AUGGUGGCAAUGUCCAAAGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCCACUUGAGGGGUGAGAG ((((..((.(((...(((----((..((((....(((....((....))...)))....))))..)))))..))).)).))))........... ( -27.60) >DroEre_CAF1 103270 89 + 1 CAGGAAAGGCGUGGGC---UUGAAGGUGGCAAUGUCCAAGGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCCACUUGAGGGGUG--AG ..........((((.(---(((((..((((....(((...((.....))...)))....))))...)))))...).))))..........--.. ( -23.40) >consensus CAGGGAGGGCGUGGGUGG_U_GAUGGUGGCAAUGUCCAAAGGGAAUGCUAAGGGAAAUGGCCAAAAUUCAAAAUGGCCACUUGAGGGGUG__AG ((((......................(((((...(((....))).))))).......((((((..........))))))))))........... (-18.02 = -18.13 + 0.11)

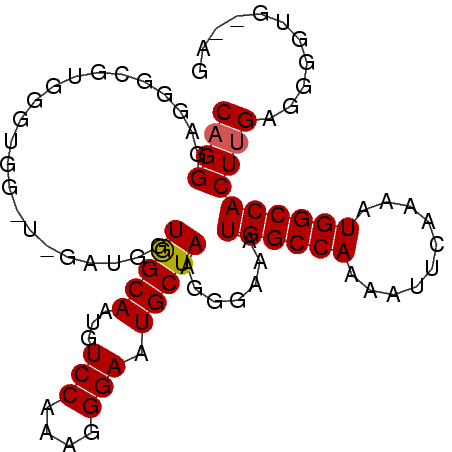

| Location | 7,109,729 – 7,109,821 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -15.73 |
| Energy contribution | -15.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7109729 92 - 22224390 CU--CACCCCUCAAGUGGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCUUUGGACAUUGCUACCAUCCAUCCACCCACGCCCUCCCAG ..--........(((((((((..........)))))))))....(((((.(((....)))...))))).......................... ( -18.20) >DroSec_CAF1 100960 90 - 1 CUCUCACCCCUCAAGUGGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCUUUGGACAUUGCCACCAU----CCACCCACGCCCUCCCUG ............(((((((((..........)))))))))......(((.(((....)))...)))......----.................. ( -15.60) >DroEre_CAF1 103270 89 - 1 CU--CACCCCUCAAGUGGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCCUUGGACAUUGCCACCUUCAA---GCCCACGCCUUUCCUG ..--..........((((....((((((...((((.((.(((...((.......)).))).)).))))..)))))---).)))).......... ( -15.70) >consensus CU__CACCCCUCAAGUGGCCAUUUUGAAUUUUGGCCAUUUCCCUUAGCAUUCCCUUUGGACAUUGCCACCAUC_A_CCACCCACGCCCUCCCUG ............(((((((((..........)))))))))......(((.(((....)))...)))............................ (-15.73 = -15.73 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:11 2006