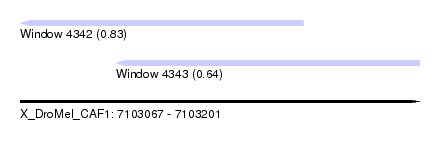
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,103,067 – 7,103,201 |
| Length | 134 |
| Max. P | 0.827447 |

| Location | 7,103,067 – 7,103,162 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 78.45 |
| Mean single sequence MFE | -9.66 |
| Consensus MFE | -8.82 |
| Energy contribution | -8.58 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7103067 95 - 22224390 AGAUGCUACGUGCUGCACACGGUCCACAUGACAUCUGCAUACAUCUCCCUCAUAUCCUCUUUCAUUUCUCCUAUUCGUUUUUUUUUUUUUUUUCA ..((((..((((.....))))(((.....)))....))))....................................................... ( -9.30) >DroPse_CAF1 49440 86 - 1 CGAUGCUACGUGAUGCACAUGGUCCACAUGACAUCUGCAUA--UUUCUCUUAUAUCUUCC----UCUCUUCUGUCU---UUUUUUUCGCUUUCUA ..((((.....((((..((((.....)))).)))).)))).--.................----............---................ ( -10.20) >DroSec_CAF1 93977 85 - 1 AGAUGCUACGUGCUGCACACGGUCCACAUGACAUCUGCAUACAUCUCCCUCAUAUCCUCUUUCAUUUCUCCUAUUC----------UUUUUUUCA ..((((..((((.....))))(((.....)))....))))....................................----------......... ( -9.30) >DroSim_CAF1 94029 85 - 1 AGAUGCUACGUGCUGCACACGGUCCACAUGACAUCUGCAUACAUCUCCCUCAUAUCCUCUUUCAUUUCUCCUAUUC----------UUUUUUUCA ..((((..((((.....))))(((.....)))....))))....................................----------......... ( -9.30) >DroPer_CAF1 49419 86 - 1 CGAUGCUACGUGAUGCACAUGGUCCACAUGACAUCUGCAUA--UUUCUCUUAUAUCUUCC----UCUCUUCUGUCU---UUUUUUUCGCUUUCUA ..((((.....((((..((((.....)))).)))).)))).--.................----............---................ ( -10.20) >consensus AGAUGCUACGUGCUGCACACGGUCCACAUGACAUCUGCAUACAUCUCCCUCAUAUCCUCUUUCAUUUCUCCUAUUC___UUUUUUUUUUUUUUCA ..((((..((((.....))))(((.....)))....))))....................................................... ( -8.82 = -8.58 + -0.24)



| Location | 7,103,099 – 7,103,201 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.99 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -23.89 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7103099 102 - 22224390 CAGGACUCGCAGGACUCGCCAAGGUGG-UCCACGGUCCACAGAUGCUACGUGCUGCACACGGUCCACAUGACAUC------UGCAUACAUCUCCCUCAUAUCCUCUUUC----------- ..((((.((..((((.(((....))))-))).)))))).((((((.((.((((((....)))..))).)).))))------))..........................----------- ( -28.20) >DroSec_CAF1 93999 102 - 1 CAGGACUCGCAGGACUCGCCAAGGUGG-UCCACGGUCCACAGAUGCUACGUGCUGCACACGGUCCACAUGACAUC------UGCAUACAUCUCCCUCAUAUCCUCUUUC----------- ..((((.((..((((.(((....))))-))).)))))).((((((.((.((((((....)))..))).)).))))------))..........................----------- ( -28.20) >DroSim_CAF1 94051 102 - 1 CAGGACUCGCAGGACUCGCCAAGGUGG-UCCACGGUCCACAGAUGCUACGUGCUGCACACGGUCCACAUGACAUC------UGCAUACAUCUCCCUCAUAUCCUCUUUC----------- ..((((.((..((((.(((....))))-))).)))))).((((((.((.((((((....)))..))).)).))))------))..........................----------- ( -28.20) >DroEre_CAF1 96133 113 - 1 CAGGACUCGCAGGACUCGCCAAGGUGG-UCCACGGUCCACAGAUGCUACGUGCUGCACACGGUCCACAUGACAUC------UGCAUACAUCUUCCUCACAUCCUCCUCCAUUUCUCCACA ..((((.((..((((.(((....))))-))).)))))).((((((.((.((((((....)))..))).)).))))------))..................................... ( -28.20) >DroYak_CAF1 96659 115 - 1 CAGGACUCGCAGGACUCGCCAAGGUGG-UCCACGGUCCACAGAUGCUACGUGCUGCACACGGUCCACAUGACAUCUGCAUCUGCAUACAUCUUCCUCAUAUCCU--UACAU-UCUCUAC- ..((((.((..((((.(((....))))-))).)))))).(((((((..((((.....))))(((.....)))....))))))).....................--.....-.......- ( -34.40) >DroAna_CAF1 143370 107 - 1 AAGGAUACGAAGGACUCGCCAAGGUGGGCCCACCGUCCACAGAUGCUACGUGAUGCACACGGUCCACAUGACAUC------UGCAUA--UUUCCCUCAUAUCCUUCG-UAU-CCUUU--- ((((((((((((((.........((((((.....))))))..((((..((((.....))))(((.....)))...------.)))).--...........)))))))-)))-)))).--- ( -40.00) >consensus CAGGACUCGCAGGACUCGCCAAGGUGG_UCCACGGUCCACAGAUGCUACGUGCUGCACACGGUCCACAUGACAUC______UGCAUACAUCUCCCUCAUAUCCUCUUUC___________ ..((((.((..(((.((((....)))).))).))))))....((((..((((.....))))(((.....)))..........)))).................................. (-23.89 = -24.08 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:07 2006