
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,084,773 – 7,084,915 |
| Length | 142 |
| Max. P | 0.730926 |

| Location | 7,084,773 – 7,084,881 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -51.60 |
| Consensus MFE | -28.55 |
| Energy contribution | -28.66 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
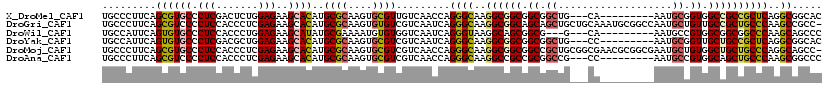
>X_DroMel_CAF1 7084773 108 + 22224390 UGCCCUUCAGCGUGCCCUCGACUCUGGAGAAGCACAUGCGCAAGUGCGUUGUCAACCAGGGCAAGGCGGCGGCGGCUG---CA---------AAUGCGGUGGCCGCCGCUCAGGCGGCAC (((((((.(((..(((...(.((((((....(((.(((((....))))))))...)))))))..)))((((((.((((---(.---------...))))).))))))))).))).)))). ( -55.20) >DroGri_CAF1 173272 119 + 1 UGCCCUUCAGCGUCCCCUCCACCCUCGAGAAGCACAUGCGCAAGUGUGUCGUCAAUCAGGGCAAGGCGGCAGCAGCUGCUGCAAAUGCGGCCAAUGCUGUUGCCGCUGCCCAAGCCGCC- .........(((..............((...((((((((....)))))).))...)).(((((..(((((((((((.(((((....)))))....))))))))))))))))....))).- ( -53.40) >DroWil_CAF1 168047 105 + 1 UGCCAUUCAGUGUGCCCUCCACCCUGGAGAAGCAUAUGCGAAAAUGUGUGGUCAAUCAGGGUAAGGCAGCGGCG---G---CA---------AAUGCCGUGGCGGCGGCCCAAGCAGCCC .(((........((((....(((((((.((.((((((......))))))..))..)))))))..))))((.(((---(---(.---------...))))).)))))(((.......))). ( -44.30) >DroYak_CAF1 77262 108 + 1 UGCCAUUCAGUGUGCCCUCGACGCUGGAGAAGCACAUGCGCAAGUGCGUCGUCAAUCAGGGCAAGGCGGCGGCGGCUG---CC---------AAUGCGGUUGCUGCCGCUCAGGCGGCAC ((((........((((((((((((((........)).((....))))))))......)))))).((((((((((((((---(.---------...))))))))))))))).....)))). ( -54.90) >DroMoj_CAF1 177633 119 + 1 UGCCCUUCAGCGUGCCCUCCACCCUCGAGAAGCACAUGCGCAAGUGCGUCGUCAACCAGGGCAAGGCGGCGGCCGCUGCGGCGAACGCGGCGAAUGCUGUGGCUGCUGCCCAGGCAGCC- ((((((.....((((.(((.......)))..))))..((((....))))........))))))..((((((..(((((((.....)))))))..))))))(((((((.....)))))))- ( -55.30) >DroAna_CAF1 118131 108 + 1 UGCCCUUCAGCGUCCCCUCCACCCUCGAGAAGCACAUGCGCAAGUGCGUCGUCAACCAGGGCAAGGCCGCCGCGGCCG---CC---------AAUGCCGUGGCAGCUGCCCAAGCGGCCC ((((((...(((.(..(((.......)))..((((........))))).))).....)))))).((((((((((((..---..---------...)))))))).(((.....))))))). ( -46.50) >consensus UGCCCUUCAGCGUGCCCUCCACCCUCGAGAAGCACAUGCGCAAGUGCGUCGUCAACCAGGGCAAGGCGGCGGCGGCUG___CA_________AAUGCCGUGGCCGCUGCCCAAGCGGCCC .........((((((.(((.......)))..))))..((((....)))).........((((..((((((.((.((...................)).)).))))))))))..))..... (-28.55 = -28.66 + 0.11)
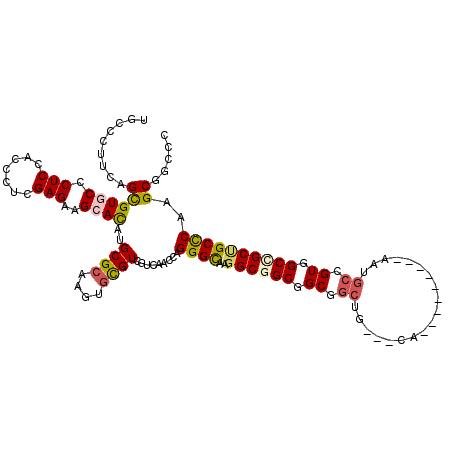
| Location | 7,084,813 – 7,084,915 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -52.08 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.77 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
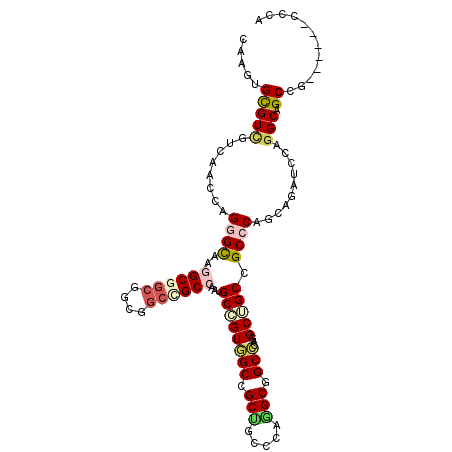
>X_DroMel_CAF1 7084813 102 + 22224390 CAAGUGCGUUGUCAACCAGGGCAAGGCGGCGGCGGCUGCAAAUGCGGUGGCCGCCGCUCAGGCGGCACAGGCUGCAGCGCAGCACAUCCAGGCCGCUG------CCCA ..................((((..(((((((((.(((((....))))).)))))))))..((((((....(((((...)))))........)))))))------))). ( -57.10) >DroPse_CAF1 26310 102 + 1 GAAGUGUGUCGUGAACCAGGGCAAGGCGGCAGCCGCCGCCAAUGCCGUGGCCGCUGCCCAGGCCGCCCAAGCCGCCGCCCAGCAGCUCCAGGCAGCCG------CCCA ..................((((..((((((.(((((.((....)).))))).))))))..(((.(((..(((.((......)).)))...))).))))------))). ( -48.30) >DroWil_CAF1 168087 99 + 1 AAAAUGUGUGGUCAAUCAGGGUAAGGCAGCGGCG---GCAAAUGCCGUGGCGGCGGCCCAAGCAGCCCAGGCGGCUGCCCAACAGAUCCAGGCGGCGG------CCCA ......((.(((......((((...((.((.(((---((....))))).)).)).)))).....)))))(((.((((((...........)))))).)------)).. ( -43.20) >DroYak_CAF1 77302 102 + 1 CAAGUGCGUCGUCAAUCAGGGCAAGGCGGCGGCGGCUGCCAAUGCGGUUGCUGCCGCUCAGGCGGCACAGGCUGCAGCGCAACACAUCCAGGCCGCCG------CCCA ..................((((..(((((((((((((((....)))))))))))))))..((((((...((.((..........)).))..)))))))------))). ( -52.60) >DroAna_CAF1 118171 108 + 1 CAAGUGCGUCGUCAACCAGGGCAAGGCCGCCGCGGCCGCCAAUGCCGUGGCAGCUGCCCAAGCGGCCCAGGCGGCCGCCCAGCAACUGCAGGCGGCGGCAGCCGUCCA ..................((((..(((.((((((((((((...((((((((....)))...)))))...)))))))(((..((....)).))).))))).))))))). ( -58.40) >DroPer_CAF1 24247 102 + 1 GAAGUGUGUCGUGAACCAGGGCAAGGCGGCAGCCGCCGCCAAUGCCGUGGCCGCUGCCCAGGCCGCCCAAGCUGCCGCCCAGCAGCUCCAGGCAGCCG------CCCA ..................((((..((((((.(((((.((....)).))))).))))))..(((.(((..((((((......))))))...))).))))------))). ( -52.90) >consensus CAAGUGCGUCGUCAACCAGGGCAAGGCGGCGGCGGCCGCCAAUGCCGUGGCCGCUGCCCAGGCGGCCCAGGCUGCCGCCCAGCAGAUCCAGGCAGCCG______CCCA .....(((((........((((..((((((....))))))...((((((((.(((.....))).)))...))))).))))..........))).))............ (-27.93 = -28.77 + 0.84)

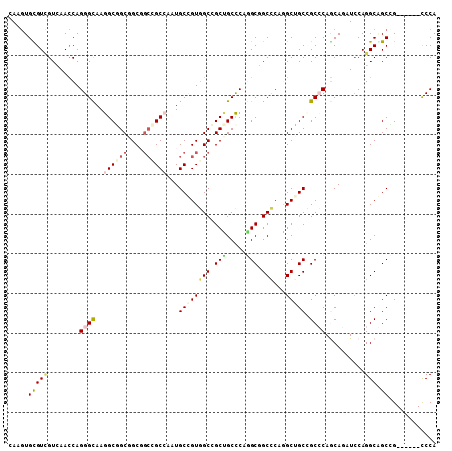
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:04 2006