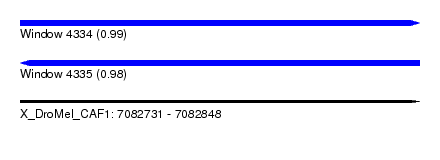
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,082,731 – 7,082,848 |
| Length | 117 |
| Max. P | 0.985224 |

| Location | 7,082,731 – 7,082,848 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -50.53 |
| Consensus MFE | -29.50 |
| Energy contribution | -32.46 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7082731 117 + 22224390 GCAGCAGCAACAAAGAGAAAAUCUGGCUUCGGCCAUGGCCGCUGGCAUGCG---UCAUCAUCCACUGUUGCCGCCGCCAGAGGCUAUGCACGCCAAUCCCUUCCAGCUGCUGCGCAUGCC ((((((((.....(((.....)))(((....((.((((((.(((((..(((---.((.((.....)).)).))).))))).))))))))..)))...........))))))))....... ( -49.30) >DroPse_CAF1 24114 117 + 1 GCAGCAGCAGCAGCGCGAGAACCUCGCUUCGGCAAUGGCCGCCGGAAUGCG---GCAUCAUCCGCUGCUGGCGCCGCCAGAGGCGAUGCAUGCCAAUCCCUUCCAGCUGCUGCGCAUGCC (((((.(((((((((((((...)))))((((((.......))))))....(---((((((((.(((.(((((...))))).))))))).)))))...........)))))))))).))). ( -60.40) >DroWil_CAF1 166005 111 + 1 CCAACAAC---------AAAACUUGGCCUCGGCCAUGGCCGCUGGCAUGAGGCAUCAUCAUCCAUUGUUGCCCCCACCAGAGGCCAUGCAUGCCAAUCCCUUCCAGCUGCUGCGUAUGCC ........---------.......(((....)))((((((.((((...(.((((.((........)).)))))...)))).))))))((((((((...............)).)))))). ( -35.96) >DroYak_CAF1 75196 117 + 1 GCAGCAGCAACAGAGAGAGAAUCUGGCAUCAGCCAUGGCCGCUGGCAUGCG---UCAUCAUCCGCUGCUGCCGCCGCCAGAGGCCAUGCACGCCAAUCCCUUCCAGCUGCUGCGCAUGCC ((((((((....(..((.((...((((....((.((((((.(((((..(((---.((.((.....)).)).))).))))).))))))))..)))).)).))..).))))))))....... ( -50.80) >DroAna_CAF1 116110 117 + 1 GCAGCAGCAACAGCGCGAGAGCCUGGCCACUGCCAUGGCGGCUGGGAUGCG---GCACCAUCCGCUCCUGCCGCCACCGGAGGCCAUGCACCCGAAUCCCUUCCAGCUGCUGAGGAUGCC .(((((((....((((....)).(((((.(((...(((((((.((((.(((---(......))))))))))))))).))).))))).))....(((....)))..)))))))........ ( -56.20) >consensus GCAGCAGCAACAGAGAGAGAACCUGGCCUCGGCCAUGGCCGCUGGCAUGCG___UCAUCAUCCGCUGCUGCCGCCGCCAGAGGCCAUGCACGCCAAUCCCUUCCAGCUGCUGCGCAUGCC ((((((((................(((....)))((((((.((((...(((....((........))....)))..)))).))))))..................))))))))....... (-29.50 = -32.46 + 2.96)


| Location | 7,082,731 – 7,082,848 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -58.00 |
| Consensus MFE | -36.10 |
| Energy contribution | -37.62 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7082731 117 - 22224390 GGCAUGCGCAGCAGCUGGAAGGGAUUGGCGUGCAUAGCCUCUGGCGGCGGCAACAGUGGAUGAUGA---CGCAUGCCAGCGGCCAUGGCCGAAGCCAGAUUUUCUCUUUGUUGCUGCUGC .......((((((((..(((((((..(((.......)))((((((((((....).(((........---)))..)))..((((....))))..))))))...)))))))...)))))))) ( -55.00) >DroPse_CAF1 24114 117 - 1 GGCAUGCGCAGCAGCUGGAAGGGAUUGGCAUGCAUCGCCUCUGGCGGCGCCAGCAGCGGAUGAUGC---CGCAUUCCGGCGGCCAUUGCCGAAGCGAGGUUCUCGCGCUGCUGCUGCUGC .(((.((((((((((.....(((((((((((.((((((..(((((...)))))..)).))))))))---)).)))))..((((....))))..(((((...))))))))))))).))))) ( -61.20) >DroWil_CAF1 166005 111 - 1 GGCAUACGCAGCAGCUGGAAGGGAUUGGCAUGCAUGGCCUCUGGUGGGGGCAACAAUGGAUGAUGAUGCCUCAUGCCAGCGGCCAUGGCCGAGGCCAAGUUUU---------GUUGUUGG ........(((((((..((..((.(((((...(((((((.((((((((((((.((........)).)))))).)))))).))))))))))))..))...))..---------))))))). ( -51.90) >DroYak_CAF1 75196 117 - 1 GGCAUGCGCAGCAGCUGGAAGGGAUUGGCGUGCAUGGCCUCUGGCGGCGGCAGCAGCGGAUGAUGA---CGCAUGCCAGCGGCCAUGGCUGAUGCCAGAUUCUCUCUCUGUUGCUGCUGC .....((((((((((.(..((((((((((((((((((((.(((((((((.((.((.....)).)).---))).)))))).)))))).))..)))))))..)))))..).)))))))).)) ( -61.60) >DroAna_CAF1 116110 117 - 1 GGCAUCCUCAGCAGCUGGAAGGGAUUCGGGUGCAUGGCCUCCGGUGGCGGCAGGAGCGGAUGGUGC---CGCAUCCCAGCCGCCAUGGCAGUGGCCAGGCUCUCGCGCUGUUGCUGCUGC ........(((((((....((.(...((((.((.(((((..(.((((((((.(((((((......)---))).)))..)))))))).)....))))).)).))))).))...))))))). ( -60.30) >consensus GGCAUGCGCAGCAGCUGGAAGGGAUUGGCGUGCAUGGCCUCUGGCGGCGGCAGCAGCGGAUGAUGA___CGCAUGCCAGCGGCCAUGGCCGAAGCCAGGUUCUCGCGCUGUUGCUGCUGC .......((((((((.(..((((.(((((..((((((((.(((((((((.((.((.....)).))....))).)))))).)))))).))....))))).))))..)...))))))))... (-36.10 = -37.62 + 1.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:00 2006