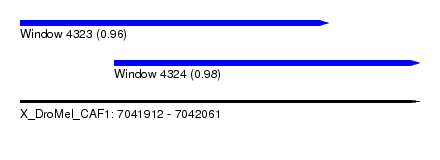
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,041,912 – 7,042,061 |
| Length | 149 |
| Max. P | 0.977714 |

| Location | 7,041,912 – 7,042,027 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.31 |
| Mean single sequence MFE | -18.27 |
| Consensus MFE | -18.01 |
| Energy contribution | -18.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7041912 115 + 22224390 AUUUUUUUCAAGUGUCUCCGGCGUGUGUUAUCAACAAAAAUAAAAUACGAAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCU ...................(((((((((((((((..............................)))))..))))))).)))...(((((((.....)))))))........... ( -18.01) >DroSec_CAF1 33353 106 + 1 AUUUUUUUCAAGUGUCUCCGGCGUGUGUUAUCAACAAAA--A-------AAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCU ...................(((((((((((((((.....--.-------...............)))))..))))))).)))...(((((((.....)))))))........... ( -18.40) >DroSim_CAF1 33365 108 + 1 AUUUUUUUCAAGUGUCUCCGGCGUGUGUUAUCAACAAAAAAA-------AAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCU ...................(((((((((((((((........-------...............)))))..))))))).)))...(((((((.....)))))))........... ( -18.30) >DroEre_CAF1 33501 104 + 1 AUUUUUUUCAAGUGUCUCCGGCGUGUGUUAUCAACAAAAA-----------AUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCCUCUUUUCU ...................(((((((((((((((......-----------.............)))))..))))))).)))...(((((((.....)))))))........... ( -18.51) >DroYak_CAF1 32084 112 + 1 AUUUUUUUCAAGUGUCUCCGGCGUGUGUUAUCAACAAAAAAAAAAUA---UAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCC ...................(((((((((((((((.............---..............)))))..))))))).)))...(((((((.....)))))))........... ( -18.13) >consensus AUUUUUUUCAAGUGUCUCCGGCGUGUGUUAUCAACAAAAA_A_______AAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCU ...................(((((((((((((((..............................)))))..))))))).)))...(((((((.....)))))))........... (-18.01 = -18.01 + -0.00)


| Location | 7,041,947 – 7,042,061 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -17.34 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7041947 114 + 22224390 AAAAAUAAAAUACGAAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCUAUACAAAUGAAACCGCGUGCGUUUGUGACAACAA------ ................................(((..(((.((.((.((((((((((.....)))))))......((((.........))))..))).)))).)))...)))..------ ( -17.20) >DroSec_CAF1 33388 105 + 1 AAAA--A-------AAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCUAUAUAAAUGAAACCGCGUGCGUUUGUGACAACAA------ ....--.-------..................(((..(((.((.((.((((((((((.....)))))))......((((.........))))..))).)))).)))...)))..------ ( -17.20) >DroSim_CAF1 33400 107 + 1 AAAAAAA-------AAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCUAUACAAAUGAAACCGCGUGCGUUUGUGACAACAA------ .......-------..................(((..(((.((.((.((((((((((.....)))))))......((((.........))))..))).)))).)))...)))..------ ( -17.20) >DroEre_CAF1 33536 109 + 1 AAAAA-----------AUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCCUCUUUUCUAUACAAAUGAAACCGCGUGCGUUUGUGACAACAACAACAA .....-----------.............(((.....(((.((.((.((((((((((.....)))))))......((((.........))))..))).)))).))).....)))...... ( -17.30) >DroYak_CAF1 32119 117 + 1 AAAAAAAAAAUA---UAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCCAUACAAAUGAAACCGCGUGCGUUUGUGACAACAACACCAA ............---..............(((((..........((.((((((((((.....)))))))..........(((....))).....))).))(((.((....)))))))))) ( -17.80) >consensus AAAAA_A_______AAAUACAAAAUCCAAUUGGUUAAACAUACAGCCUGCGAAAACCCAAUUGGUUUUCCCUUCUUUUCUAUACAAAUGAAACCGCGUGCGUUUGUGACAACAA______ ................................(((..(((.((.((.((((((((((.....)))))))......((((.........))))..))).)))).)))...)))........ (-16.96 = -16.96 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:50 2006