
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,032,121 – 7,032,212 |
| Length | 91 |
| Max. P | 0.583619 |

| Location | 7,032,121 – 7,032,212 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -20.88 |
| Energy contribution | -22.22 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
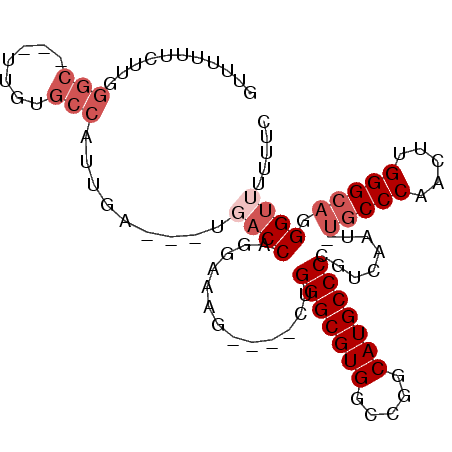
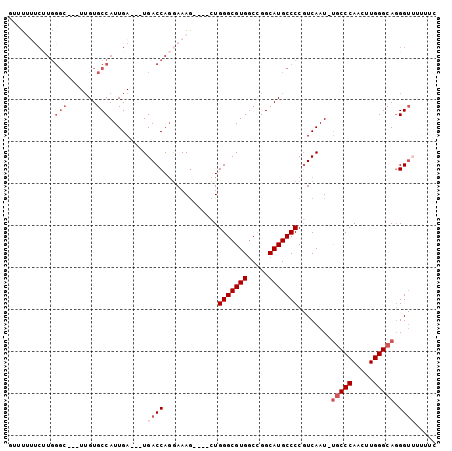
>X_DroMel_CAF1 7032121 91 + 22224390 GUUUUUUCUUGGGC---UUGUGCCAUUGA---UGACCAGGAAAG----CUGGGCGUGGCCGGCAUGCCCCGUCAAU-UGCCCAACUUGGGCAGGGUUUUUUC (((...((...(((---....)))...))---.)))..((((((----(((((((((.....)))))))......(-(((((.....)))))))))))))). ( -32.60) >DroVir_CAF1 104695 87 + 1 GCCUGGGCCUGUGCCUGUGGUGCCAUUGA---UGACCACAAAAA----CUGGGCGUGGCUGGCAUGCCCC-UCAAUUCACCCAACUUGGGCAUGG------- (((..((...(((..((((((........---..))))))....----..(((((((.....))))))).-......)))....))..)))....------- ( -29.50) >DroSec_CAF1 23623 90 + 1 UUUUUUUCUUGGGC---UUGUGCCAUUGA---UGACCAGGAAAG----CUGGGCGUG-CCGGCAUGCCCCGUCAAU-UGCCCAACUUGGGCAGGGUUUUUUC ......((...(((---....)))...))---.((((......(----(.(((((((-....))))))).))...(-(((((.....))))))))))..... ( -32.50) >DroEre_CAF1 23674 91 + 1 GUUUUUUCUUGGGC---UUGUGCCAUUGA---UGACCAGGAAAG----CUGGGCGUGGCCGGCAUGCCCCGUCAAU-UGCCCAACUUGGGCAGGGUUUUUUU (((...((...(((---....)))...))---.))).(((((((----(((((((((.....)))))))......(-(((((.....))))))))))))))) ( -33.10) >DroYak_CAF1 21735 90 + 1 -UUUUUUCUUGGGC---UUGUGCCAUUGA---UGACCAGGAAAG----CUGGGCGUGGCCGGCAUGCCCCGUCAAU-UGCCCAACUUGGGCAGGGUUUUUUC -.....((...(((---....)))...))---.((((......(----(.(((((((.....))))))).))...(-(((((.....))))))))))..... ( -31.50) >DroAna_CAF1 68109 81 + 1 -----------GCG---UUGUGCCAUUGAUGAUGACCAGGAAAGAGUCAUGGGCGUGGCCGGCAUGCCCCGUCAAU-UGCCCAACUUGGGCAGGGU------ -----------...---....(((.((((((((((((......).)))))(((((((.....)))))))))))))(-(((((.....)))))))))------ ( -33.00) >consensus GUUUUUUCUUGGGC___UUGUGCCAUUGA___UGACCAGGAAAG____CUGGGCGUGGCCGGCAUGCCCCGUCAAU_UGCCCAACUUGGGCAGGGUUUUUUC ...........(((.......))).........((((.............(((((((.....)))))))........(((((.....))))).))))..... (-20.88 = -22.22 + 1.33)

| Location | 7,032,121 – 7,032,212 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.27 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
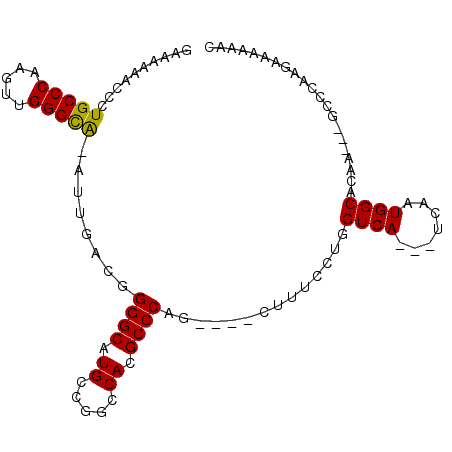
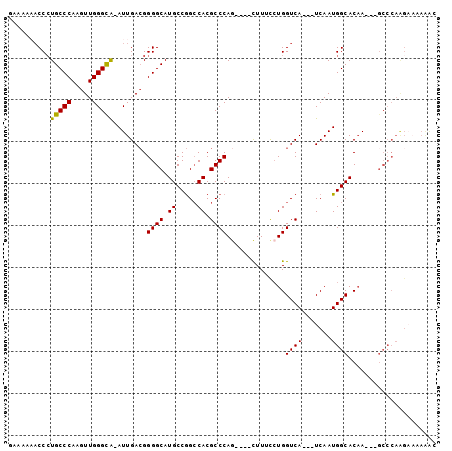
>X_DroMel_CAF1 7032121 91 - 22224390 GAAAAAACCCUGCCCAAGUUGGGCA-AUUGACGGGGCAUGCCGGCCACGCCCAG----CUUUCCUGGUCA---UCAAUGGCACAA---GCCCAAGAAAAAAC ..........(((((.....)))))-.......((((.(((((.......((((----.....))))...---....)))))...---)))).......... ( -27.34) >DroVir_CAF1 104695 87 - 1 -------CCAUGCCCAAGUUGGGUGAAUUGA-GGGGCAUGCCAGCCACGCCCAG----UUUUUGUGGUCA---UCAAUGGCACCACAGGCACAGGCCCAGGC -------.(((((((.((((.....))))..-.)))))))...(((..(((..(----(.(((((((((.---......).)))))))).)).)))...))) ( -29.60) >DroSec_CAF1 23623 90 - 1 GAAAAAACCCUGCCCAAGUUGGGCA-AUUGACGGGGCAUGCCGG-CACGCCCAG----CUUUCCUGGUCA---UCAAUGGCACAA---GCCCAAGAAAAAAA ..........(((((.....)))))-.......((((.(((((.-.....((((----.....))))...---....)))))...---)))).......... ( -26.92) >DroEre_CAF1 23674 91 - 1 AAAAAAACCCUGCCCAAGUUGGGCA-AUUGACGGGGCAUGCCGGCCACGCCCAG----CUUUCCUGGUCA---UCAAUGGCACAA---GCCCAAGAAAAAAC ..........(((((.....)))))-.......((((.(((((.......((((----.....))))...---....)))))...---)))).......... ( -27.34) >DroYak_CAF1 21735 90 - 1 GAAAAAACCCUGCCCAAGUUGGGCA-AUUGACGGGGCAUGCCGGCCACGCCCAG----CUUUCCUGGUCA---UCAAUGGCACAA---GCCCAAGAAAAAA- ..........(((((.....)))))-.......((((.(((((.......((((----.....))))...---....)))))...---)))).........- ( -27.34) >DroAna_CAF1 68109 81 - 1 ------ACCCUGCCCAAGUUGGGCA-AUUGACGGGGCAUGCCGGCCACGCCCAUGACUCUUUCCUGGUCAUCAUCAAUGGCACAA---CGC----------- ------.((((((((.....)))))-......)))...(((((((...))).((((((.......)))))).......))))...---...----------- ( -24.00) >consensus GAAAAAACCCUGCCCAAGUUGGGCA_AUUGACGGGGCAUGCCGGCCACGCCCAG____CUUUCCUGGUCA___UCAAUGGCACAA___GCCCAAGAAAAAAC ..........(((((.....)))))........((((.((.....)).))))..............((((.......))))..................... (-17.54 = -17.27 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:43 2006