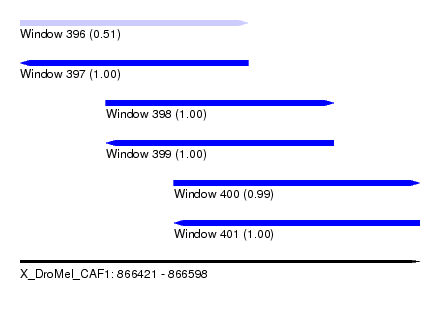
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 866,421 – 866,598 |
| Length | 177 |
| Max. P | 0.999999 |

| Location | 866,421 – 866,522 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.16 |
| Mean single sequence MFE | -18.41 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.47 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866421 101 + 22224390 ACGCACACAGCGAAAUUUUCGUUUUGAGUGAGUUUUU--U-GCAUUCCUACUGAGUUUUACUCUGAUUUUA---------------UUCUGCACAAUAAACUCACUCAC-UUGAGUUUGA .(((.....))).....((((...(((((((((((..--(-(((........(((.....))).((.....---------------.))))))....))))))))))).-.))))..... ( -25.20) >DroEre_CAF1 72870 117 + 1 ACGC---CAGCGAAAUAUUUGUUUUGAGUGAGUUUUUGUUAGAUAUUAUACUGAGUUUUACUCUAUUUUUUAUUUCUUUGUUUUUUUUUUGCACCAACAACCCACUCACAUUGAGUUUGA ..((---(((((((((....)))))).((((((..((((((((((.......(((.....))).......)))))...(((.........)))..)))))...)))))).))).)).... ( -15.54) >DroYak_CAF1 69077 94 + 1 ACGC---CAGCGAAGUAUUUGUUUUGAGUGAGUUUUU--UCACUAUUCAACUGAGUUUUACUCUAUUUUUAAUUUUUU---------------------ACUUACUCACUUUGAGUUUGA .((.---...))(((((...(((...((((((....)--)))))....))).(((.....)))..............)---------------------))))((((.....)))).... ( -14.50) >consensus ACGC___CAGCGAAAUAUUUGUUUUGAGUGAGUUUUU__U_GCUAUUCUACUGAGUUUUACUCUAUUUUUAAUUU_UU________UU_UGCAC_A__AACUCACUCAC_UUGAGUUUGA .(((.....))).....((((...((((((((((..................(((.....)))((....))...........................))))))))))...))))..... (-11.13 = -11.47 + 0.34)

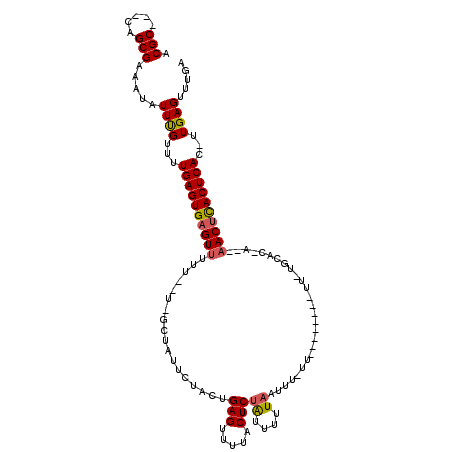
| Location | 866,421 – 866,522 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.16 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 5.64 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866421 101 - 22224390 UCAAACUCAA-GUGAGUGAGUUUAUUGUGCAGAA---------------UAAAAUCAGAGUAAAACUCAGUAGGAAUGC-A--AAAAACUCACUCAAAACGAAAAUUUCGCUGUGUGCGU ..........-.(((((((((((....((((((.---------------.....)).(((.....)))........)))-)--..)))))))))))............(((.....))). ( -25.00) >DroEre_CAF1 72870 117 - 1 UCAAACUCAAUGUGAGUGGGUUGUUGGUGCAAAAAAAAAACAAAGAAAUAAAAAAUAGAGUAAAACUCAGUAUAAUAUCUAACAAAAACUCACUCAAAACAAAUAUUUCGCUG---GCGU ............(((((((((((((((................(....)........(((.....)))..........))))))...))))))))).................---.... ( -18.10) >DroYak_CAF1 69077 94 - 1 UCAAACUCAAAGUGAGUAAGU---------------------AAAAAAUUAAAAAUAGAGUAAAACUCAGUUGAAUAGUGA--AAAAACUCACUCAAAACAAAUACUUCGCUG---GCGU ...........((.(((((((---------------------(..............(((.....))).(((....(((((--......)))))...)))...))))).))).---)).. ( -17.70) >consensus UCAAACUCAA_GUGAGUGAGUU__U_GUGCA_AA________AA_AAAUUAAAAAUAGAGUAAAACUCAGUAGAAUAGC_A__AAAAACUCACUCAAAACAAAUAUUUCGCUG___GCGU ............((((((((((...................................(((.....))).(........).......))))))))))............(((.....))). (-13.37 = -13.60 + 0.23)

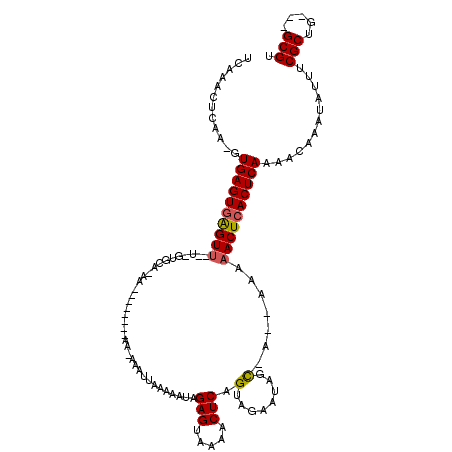
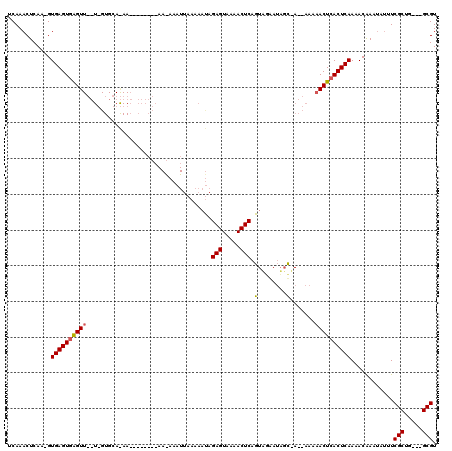
| Location | 866,459 – 866,560 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.74 |
| Mean single sequence MFE | -17.40 |
| Consensus MFE | -13.88 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.80 |
| SVM decision value | 6.16 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866459 101 + 22224390 -GCAUUCCUACUGAGUUUUACUCUGAUUUUA---------------UUCUGCACAAUAAACUCACUCAC-UUGAGUUUGAGUAAGUCCCAAA-AUUUCCCCUUCUU-UUAGGACAAUUUU -(((........(((.....))).((.....---------------.)))))....((((((((.....-.)))))))).....((((.(((-(..........))-)).))))...... ( -16.30) >DroEre_CAF1 72907 120 + 1 AGAUAUUAUACUGAGUUUUACUCUAUUUUUUAUUUCUUUGUUUUUUUUUUGCACCAACAACCCACUCACAUUGAGUUUGAGUACGUCCCAAAAAUUUCCCCUCCCUCUUGGGACAAUUUU ..................(((((..............(((((.............)))))...((((.....))))..))))).(((((((................)))))))...... ( -16.91) >DroYak_CAF1 69112 99 + 1 CACUAUUCAACUGAGUUUUACUCUAUUUUUAAUUUUUU---------------------ACUUACUCACUUUGAGUUUGAGUAAGUCCCAAAAAUUUCCCCUCCUUCUUGGGACAAUUUU ...(((((((..(((.....)))...............---------------------....((((.....))))))))))).(((((((................)))))))...... ( -18.99) >consensus _GCUAUUCUACUGAGUUUUACUCUAUUUUUAAUUU_UU________UU_UGCAC_A__AACUCACUCAC_UUGAGUUUGAGUAAGUCCCAAAAAUUUCCCCUCCUUCUUGGGACAAUUUU ............(((.....)))....................................((((((((.....))))..))))..(((((((................)))))))...... (-13.88 = -14.32 + 0.45)

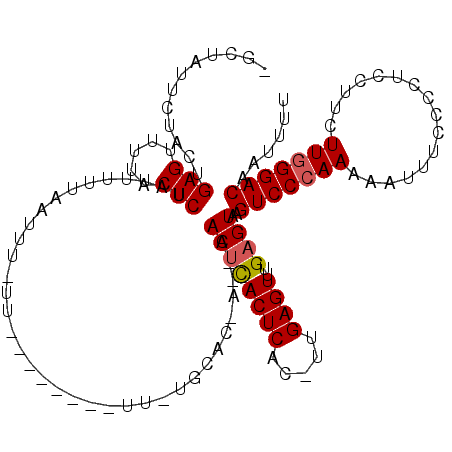
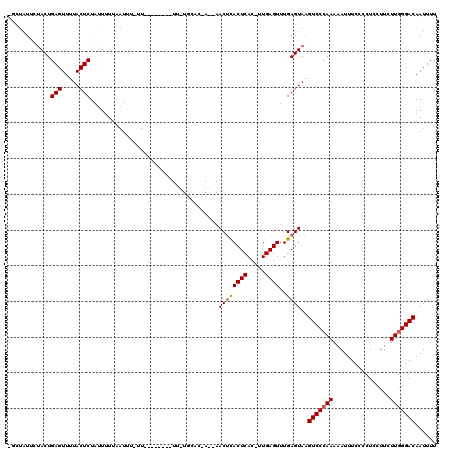
| Location | 866,459 – 866,560 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.74 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.80 |
| SVM decision value | 6.54 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866459 101 - 22224390 AAAAUUGUCCUAA-AAGAAGGGGAAAU-UUUGGGACUUACUCAAACUCAA-GUGAGUGAGUUUAUUGUGCAGAA---------------UAAAAUCAGAGUAAAACUCAGUAGGAAUGC- ......(((((((-((..........)-))))))))((((((..((((..-..))))((.((((((......))---------------)))).)).))))))................- ( -22.30) >DroEre_CAF1 72907 120 - 1 AAAAUUGUCCCAAGAGGGAGGGGAAAUUUUUGGGACGUACUCAAACUCAAUGUGAGUGGGUUGUUGGUGCAAAAAAAAAACAAAGAAAUAAAAAAUAGAGUAAAACUCAGUAUAAUAUCU ......((((((((((..........))))))))))(((((..((((((.......))))))...)))))...........................(((.....)))............ ( -25.90) >DroYak_CAF1 69112 99 - 1 AAAAUUGUCCCAAGAAGGAGGGGAAAUUUUUGGGACUUACUCAAACUCAAAGUGAGUAAGU---------------------AAAAAAUUAAAAAUAGAGUAAAACUCAGUUGAAUAGUG ......((((((((((..........))))))))))(((((...((((.....)))).)))---------------------))...((((..(((.(((.....))).)))...)))). ( -21.70) >consensus AAAAUUGUCCCAAGAAGGAGGGGAAAUUUUUGGGACUUACUCAAACUCAA_GUGAGUGAGUU__U_GUGCA_AA________AA_AAAUUAAAAAUAGAGUAAAACUCAGUAGAAUAGC_ ......((((((((((..........))))))))))(((((...((((.....))))........................................(((.....))))))))....... (-18.65 = -18.77 + 0.12)

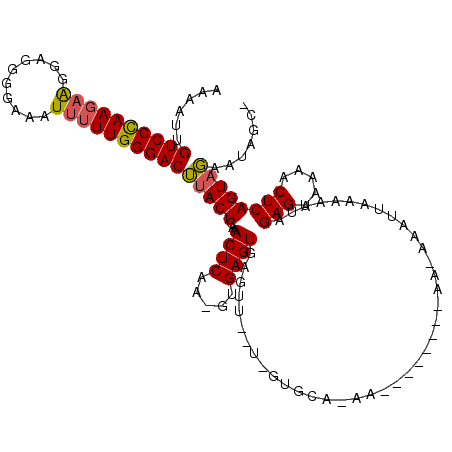
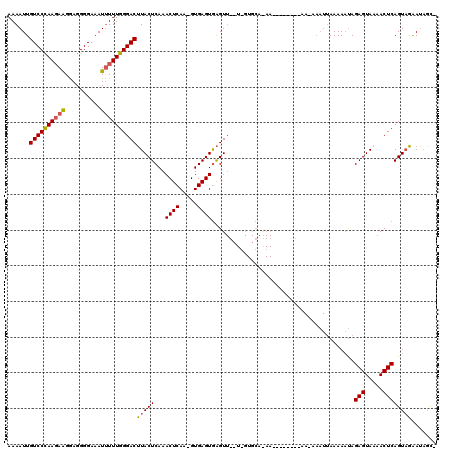
| Location | 866,489 – 866,598 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866489 109 + 22224390 ------UUCUGCACAAUAAACUCACUCAC-UUGAGUUUGAGUAAGUCCCAAA-AUUUCCCCUUCUU-UUAGGACAAUUUUUUUGCAGACGAAACUUAAAACGUUGGGAAAUUCCAGCU-- ------.((((((...((((((((.....-.)))))))).....((((.(((-(..........))-)).))))........)))))).............((((((....)))))).-- ( -26.30) >DroEre_CAF1 72947 120 + 1 UUUUUUUUUUGCACCAACAACCCACUCACAUUGAGUUUGAGUACGUCCCAAAAAUUUCCCCUCCCUCUUGGGACAAUUUUUUUGCAGACACAAAUUAAAACACAGGGAAAUUUCUGCUUU ..........(((....(((...((((.....)))))))............(((((((((.((((....))))...((..((((......))))..))......))))))))).)))... ( -21.30) >DroYak_CAF1 69150 99 + 1 -------------------ACUUACUCACUUUGAGUUUGAGUAAGUCCCAAAAAUUUCCCCUCCUUCUUGGGACAAUUUUUUGGCGGACGCAAAUGAAAACAUUGGGAAAUUCCUGCU-- -------------------(((((((((.........)))))))))..((..((((((((.((((....))))...(((((..((....))....)))))....))))))))..))..-- ( -23.60) >consensus ______UU_UGCAC_A__AACUCACUCAC_UUGAGUUUGAGUAAGUCCCAAAAAUUUCCCCUCCUUCUUGGGACAAUUUUUUUGCAGACGCAAAUUAAAACAUUGGGAAAUUCCUGCU__ ...................((((((((.....))))..))))..........((((((((.((((....))))(((.....)))....................))))))))........ (-14.42 = -14.53 + 0.12)

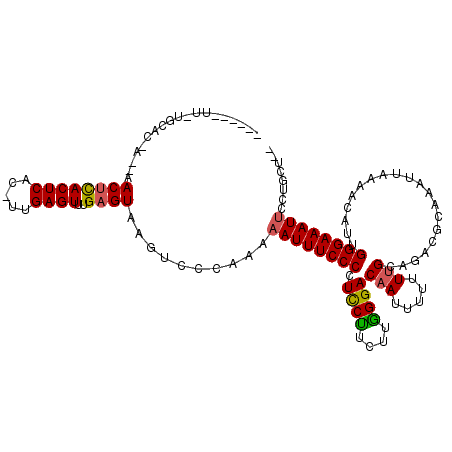
| Location | 866,489 – 866,598 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -22.08 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.79 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866489 109 - 22224390 --AGCUGGAAUUUCCCAACGUUUUAAGUUUCGUCUGCAAAAAAAUUGUCCUAA-AAGAAGGGGAAAU-UUUGGGACUUACUCAAACUCAA-GUGAGUGAGUUUAUUGUGCAGAA------ --.(.(((......))).).............((((((..((((((.((((..-......)))))))-))).(((((((((((.......-.)))))))))))....)))))).------ ( -28.40) >DroEre_CAF1 72947 120 - 1 AAAGCAGAAAUUUCCCUGUGUUUUAAUUUGUGUCUGCAAAAAAAUUGUCCCAAGAGGGAGGGGAAAUUUUUGGGACGUACUCAAACUCAAUGUGAGUGGGUUGUUGGUGCAAAAAAAAAA ....(((((((((((((.........(((((....))))).......((((....)))))))))))))))))....(((((..((((((.......))))))...))))).......... ( -35.10) >DroYak_CAF1 69150 99 - 1 --AGCAGGAAUUUCCCAAUGUUUUCAUUUGCGUCCGCCAAAAAAUUGUCCCAAGAAGGAGGGGAAAUUUUUGGGACUUACUCAAACUCAAAGUGAGUAAGU------------------- --..((((((((((((.............((....))..........(((......))).))))))))))))..(((((((((.........)))))))))------------------- ( -27.40) >consensus __AGCAGGAAUUUCCCAAUGUUUUAAUUUGCGUCUGCAAAAAAAUUGUCCCAAGAAGGAGGGGAAAUUUUUGGGACUUACUCAAACUCAA_GUGAGUGAGUU__U_GUGCA_AA______ ....((((((((((((...........((((....))))........((((....)))).)))))))))))).((((((((((.........)))))))))).................. (-22.08 = -23.87 + 1.79)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:19 2006