
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,997,344 – 6,997,584 |
| Length | 240 |
| Max. P | 0.993006 |

| Location | 6,997,344 – 6,997,464 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -31.77 |
| Energy contribution | -31.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6997344 120 + 22224390 AGAUGGGUUGGAGAUGGUGAAGUGGAGAUGGAGAUGGAGAUGAAGCUGCGAAAGCAAGCGAAAGUUCAAAAUGUGCAAAAAGCGAGCACAGAAAGCUGAAAGCACAACUCCACACACGCA ................(((..((((((.........(((.....(((((....)).))).....)))....(((((....(((...........)))....))))).)))))).)))... ( -32.90) >DroSec_CAF1 19664 112 + 1 AGAUGGGUUGGAGAUGGUGAAGUGGAGAU------GGAGAUGAAGCUGCGAAAGCAAGCGAAAGUUCAAAAUGUGCAAAAAGCGAGCACAGAAAGCUGAAAGCACAACUCCACACAC--A ................(((..((((((..------.(((.....(((((....)).))).....)))....(((((....(((...........)))....))))).)))))).)))--. ( -32.90) >DroSim_CAF1 4805 112 + 1 AGAUGGGUUGGAGAUGGUGAAGUGGAGAU------GGAGAUGAAGCUGCGAAAGCAAGCGAAAGUUCAAAAUGUGCAAAAAGCGAGCACAGAAAGCUGAAAGCACAACUCCACACAC--A ................(((..((((((..------.(((.....(((((....)).))).....)))....(((((....(((...........)))....))))).)))))).)))--. ( -32.90) >consensus AGAUGGGUUGGAGAUGGUGAAGUGGAGAU______GGAGAUGAAGCUGCGAAAGCAAGCGAAAGUUCAAAAUGUGCAAAAAGCGAGCACAGAAAGCUGAAAGCACAACUCCACACAC__A ................(((..((((((.........(((.....(((((....)).))).....)))....(((((....(((...........)))....))))).)))))).)))... (-31.77 = -31.77 + 0.00)


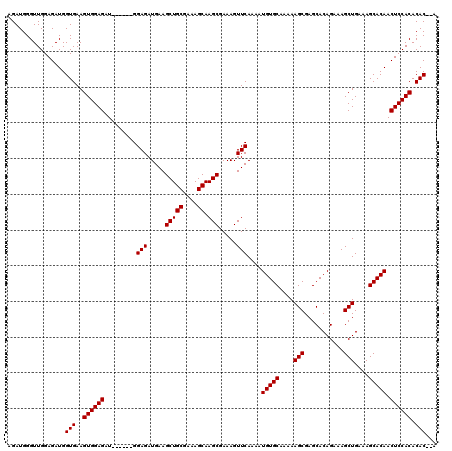
| Location | 6,997,344 – 6,997,464 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6997344 120 - 22224390 UGCGUGUGUGGAGUUGUGCUUUCAGCUUUCUGUGCUCGCUUUUUGCACAUUUUGAACUUUCGCUUGCUUUCGCAGCUUCAUCUCCAUCUCCAUCUCCACUUCACCAUCUCCAACCCAUCU ((.(((.((((((.(((((....(((...........)))....)))))............(((.((....))))).................))))))..))))).............. ( -25.60) >DroSec_CAF1 19664 112 - 1 U--GUGUGUGGAGUUGUGCUUUCAGCUUUCUGUGCUCGCUUUUUGCACAUUUUGAACUUUCGCUUGCUUUCGCAGCUUCAUCUCC------AUCUCCACUUCACCAUCUCCAACCCAUCU .--(((.((((((.(((((....(((...........)))....)))))............(((.((....))))).........------..))))))..)))................ ( -24.70) >DroSim_CAF1 4805 112 - 1 U--GUGUGUGGAGUUGUGCUUUCAGCUUUCUGUGCUCGCUUUUUGCACAUUUUGAACUUUCGCUUGCUUUCGCAGCUUCAUCUCC------AUCUCCACUUCACCAUCUCCAACCCAUCU .--(((.((((((.(((((....(((...........)))....)))))............(((.((....))))).........------..))))))..)))................ ( -24.70) >consensus U__GUGUGUGGAGUUGUGCUUUCAGCUUUCUGUGCUCGCUUUUUGCACAUUUUGAACUUUCGCUUGCUUUCGCAGCUUCAUCUCC______AUCUCCACUUCACCAUCUCCAACCCAUCU ...(((.((((((.(((((....(((...........)))....)))))............(((.((....))))).................))))))..)))................ (-24.80 = -24.80 + 0.00)

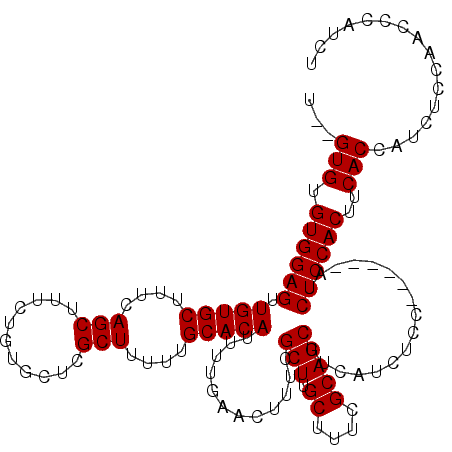

| Location | 6,997,464 – 6,997,584 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6997464 120 + 22224390 CACCCACACACCCACACAUGUGAAAGCACACAGACUACCGUGCGCCCAAAUGCGUGCGAUAUAUGCAUAUGCAUGUACAGUCCAACUUCUAUAAGCCUCACUUUUGUGCAAAAAGGAAAA ...((.((((........))))...(((((..((((..((..(((......)))..)).(((((((....))))))).))))...(((....))).........))))).....)).... ( -30.40) >DroSec_CAF1 19776 97 + 1 CAC--------CCACACAUGUGAAAGCACACAGACAACCGUGCGCCCAAAUGCGUGCGAUAUAUGCAUAUGCAUGUACAGUCCAACUUCUAUAAGCCUCACUUUU--------------- ...--------.......((((......))))(((...((..(((......)))..)).(((((((....)))))))..))).......................--------------- ( -22.30) >DroSim_CAF1 4917 105 + 1 CACCCACACACCCACACAUGUGAAAGCACACAGACAACCGUGCGCCCAAAUGCGUGCGAUAUAUGCAUAUGCAUGUACAGUCCAACUUCUAUAAGCCUCACUUUU--------------- ..................((((......))))(((...((..(((......)))..)).(((((((....)))))))..))).......................--------------- ( -22.30) >consensus CACCCACACACCCACACAUGUGAAAGCACACAGACAACCGUGCGCCCAAAUGCGUGCGAUAUAUGCAUAUGCAUGUACAGUCCAACUUCUAUAAGCCUCACUUUU_______________ ...................((((..((...........((..(((......)))..)).(((((((....))))))).................)).))))................... (-22.20 = -22.20 + -0.00)

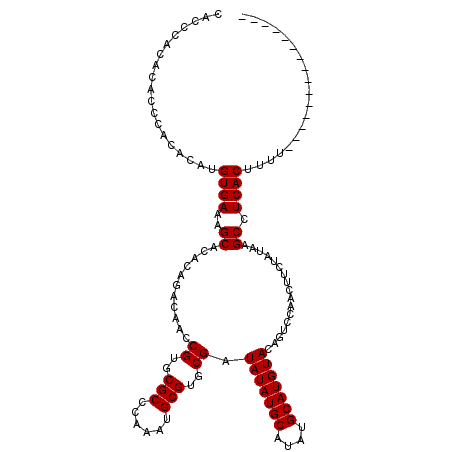
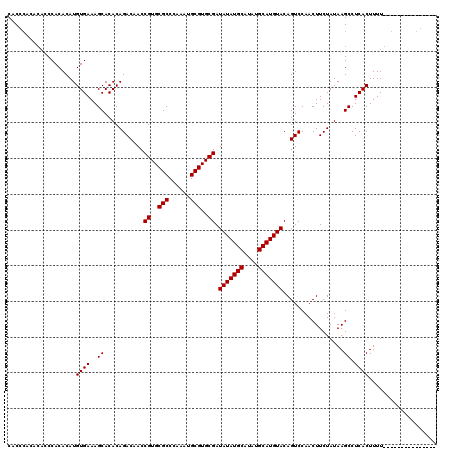
| Location | 6,997,464 – 6,997,584 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -32.30 |
| Energy contribution | -34.63 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6997464 120 - 22224390 UUUUCCUUUUUGCACAAAAGUGAGGCUUAUAGAAGUUGGACUGUACAUGCAUAUGCAUAUAUCGCACGCAUUUGGGCGCACGGUAGUCUGUGUGCUUUCACAUGUGUGGGUGUGUGGGUG ..........((((....(((..(((((....)))))..))).....))))(((.((((((((.(((((((.(((((((((((....)))))))))..)).))))))))))))))).))) ( -43.00) >DroSec_CAF1 19776 97 - 1 ---------------AAAAGUGAGGCUUAUAGAAGUUGGACUGUACAUGCAUAUGCAUAUAUCGCACGCAUUUGGGCGCACGGUUGUCUGUGUGCUUUCACAUGUGUGG--------GUG ---------------...(((..(((((....)))))..)))....((((....)))).((((.(((((((.(((((((((((....)))))))))..)).))))))))--------))) ( -32.80) >DroSim_CAF1 4917 105 - 1 ---------------AAAAGUGAGGCUUAUAGAAGUUGGACUGUACAUGCAUAUGCAUAUAUCGCACGCAUUUGGGCGCACGGUUGUCUGUGUGCUUUCACAUGUGUGGGUGUGUGGGUG ---------------...(((..(((((....)))))..))).........(((.((((((((.(((((((.(((((((((((....)))))))))..)).))))))))))))))).))) ( -38.80) >consensus _______________AAAAGUGAGGCUUAUAGAAGUUGGACUGUACAUGCAUAUGCAUAUAUCGCACGCAUUUGGGCGCACGGUUGUCUGUGUGCUUUCACAUGUGUGGGUGUGUGGGUG ..................(((..(((((....)))))..))).........(((.((((((((.(((((((.(((((((((((....)))))))))..)).))))))))))))))).))) (-32.30 = -34.63 + 2.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:12 2006