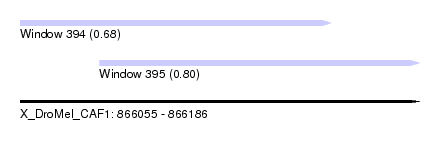
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 866,055 – 866,186 |
| Length | 131 |
| Max. P | 0.802247 |

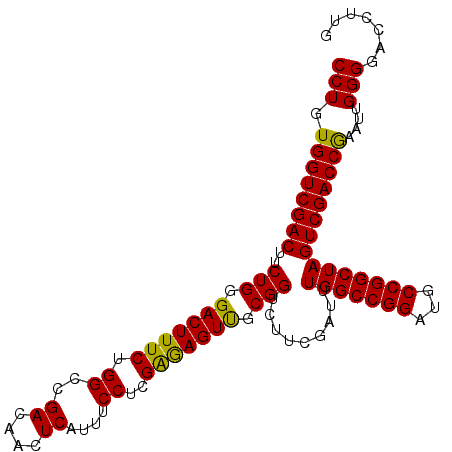
| Location | 866,055 – 866,157 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -32.91 |
| Energy contribution | -32.03 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866055 102 + 22224390 CUGCGUUUCUAGGGUCUC--------------UAGUCUGGCCUGUGGUCGACUUCUGGGACUUUCUGGCGGACAACUCAUUUCCUCGAGAGUUGCGGUCUUUGAUUUGGCCGGAUG ...((((...((((((((--------------.((((.((((...))))))))...))))))))..))))..((((((..........))))))(((((........))))).... ( -33.40) >DroEre_CAF1 72503 102 + 1 CUGCGUUUCUAGGGUCUA--------------UAGUCUGGCCUGUGGUCGACUCCUGGGACUUUCUGGCCGACAACUCAUUUCCUCGAGAGUUGCGGUAUUCGAUUUGGCCGGAUG ..................--------------..((((((((...((((((((....))........((((.((((((..........))))))))))..)))))).)))))))). ( -33.40) >DroYak_CAF1 68675 116 + 1 CUGCGUUUCUAGGGUCUGUAGUCCGUAGUCUGUAGUCUGGCCUGUGGUCGACUUCUGCGACUUUCUGGCUGACAACUCAUUUCCUCGGAAGUCGCGGUCUUCGAUUUGGCCGGAUG (((((...(((.((.(....).)).)))..)))))(((((((...((((((...(((((((((((.((.(((....)))...))..)))))))))))...)))))).))))))).. ( -48.10) >consensus CUGCGUUUCUAGGGUCUA______________UAGUCUGGCCUGUGGUCGACUUCUGGGACUUUCUGGCCGACAACUCAUUUCCUCGAGAGUUGCGGUCUUCGAUUUGGCCGGAUG ..................................((((((((...((((((...(((.(((((((.((..((....))....))..))))))).)))...)))))).)))))))). (-32.91 = -32.03 + -0.88)

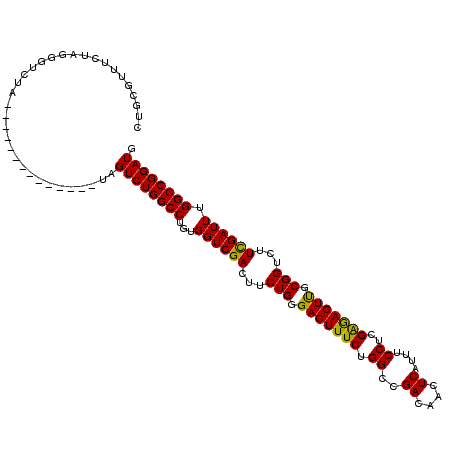
| Location | 866,081 – 866,186 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -38.45 |
| Energy contribution | -37.57 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 866081 105 + 22224390 CCUGUGGUCGACUUCUGGGACUUUCUGGCGGACAACUCAUUUCCUCGAGAGUUGCGGUCUUUGAUUUGGCCGGAUGCCGGCUAGUCGACCGAAUUGGGGACCUUG (((.((((((((..(((.(((((((.((.(((.........)))))))))))).))).........(((((((...)))))))))))))))....)))....... ( -39.50) >DroEre_CAF1 72529 105 + 1 CCUGUGGUCGACUCCUGGGACUUUCUGGCCGACAACUCAUUUCCUCGAGAGUUGCGGUAUUCGAUUUGGCCGGAUGCCGGCUAGUCGACCGAAUUGGGGACCUUG (((.(((((((((.((((.(...(((((((((((((((..........))))))((.....))..)))))))))).))))..)))))))))....)))....... ( -41.20) >DroYak_CAF1 68715 105 + 1 CCUGUGGUCGACUUCUGCGACUUUCUGGCUGACAACUCAUUUCCUCGGAAGUCGCGGUCUUCGAUUUGGCCGGAUGCCGGCUAGUCGACCAAGUUGGGGACCUUG (((.((((((((..(((((((((((.((.(((....)))...))..))))))))))).........(((((((...)))))))))))))))....)))....... ( -44.90) >consensus CCUGUGGUCGACUUCUGGGACUUUCUGGCCGACAACUCAUUUCCUCGAGAGUUGCGGUCUUCGAUUUGGCCGGAUGCCGGCUAGUCGACCGAAUUGGGGACCUUG (((.((((((((..(((.(((((((.((..((....))....))..))))))).))).........(((((((...)))))))))))))))....)))....... (-38.45 = -37.57 + -0.88)

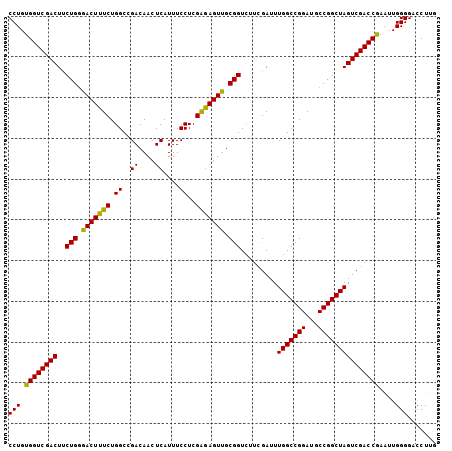
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:14 2006