
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,995,590 – 6,995,750 |
| Length | 160 |
| Max. P | 0.947757 |

| Location | 6,995,590 – 6,995,710 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6995590 120 + 22224390 UUCGCUGUUAUCCUUUUGCAACAUUGCACACAAUACAAUAUAUGCGAUUUUGGGAUCGGUGAAAGCCCACCCUUUCCUUUCUCCCAUUUCUAUUGCGUGUGCAGAUGCAUUGCUUGCAUU ...((............)).....(((((((.(((.....)))(((((..(((((..((.(((((......)))))))...))))).....))))))))))))((((((.....)))))) ( -31.20) >DroSec_CAF1 17946 120 + 1 UUCGCUGUUAUCCUUUUGCAACAUUGCACACAAUACAAUAUAUGCGAUUUUGGGAUCGCUGAAAGCCCACCCUUUCCCUUCACCCAUUUCUAUUGCGUGUGCAGAUGCAUUGCUUGCAUU ...((............)).....(((((((.(((.....)))(((((..((((......(((((......)))))......)))).....))))))))))))((((((.....)))))) ( -28.20) >DroSim_CAF1 3096 120 + 1 UUCGCUGUUAUCCUUUUGCAACAUUGCACACAAUACAAUAUAUGCGAUUUUGGGAUCGCUGAAAGCCCACCCUUUCCCUUCUCCCAUUUCUAUUGCGUGUACAGAUGCAUUGCUUGCAUU ....(((.........((((....))))..........((((((((((..(((((.....(((((......))))).....))))).....)))))))))))))(((((.....))))). ( -28.50) >consensus UUCGCUGUUAUCCUUUUGCAACAUUGCACACAAUACAAUAUAUGCGAUUUUGGGAUCGCUGAAAGCCCACCCUUUCCCUUCUCCCAUUUCUAUUGCGUGUGCAGAUGCAUUGCUUGCAUU ....((((........((((....))))...........(((((((((..(((((.....(((((......))))).....))))).....)))))))))))))(((((.....))))). (-27.42 = -27.53 + 0.11)

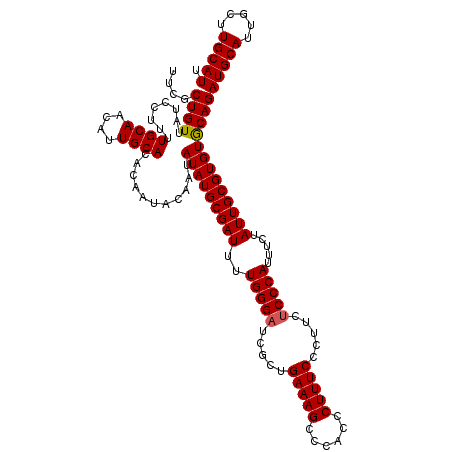
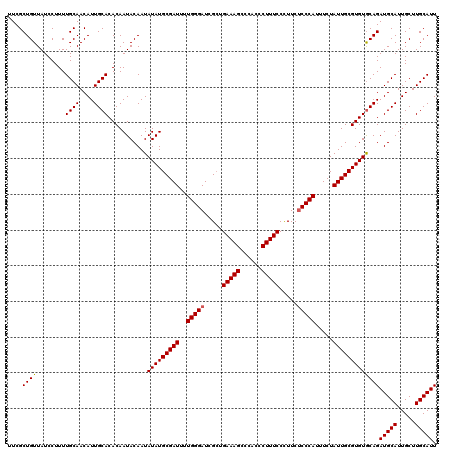
| Location | 6,995,590 – 6,995,710 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -31.05 |
| Energy contribution | -31.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6995590 120 - 22224390 AAUGCAAGCAAUGCAUCUGCACACGCAAUAGAAAUGGGAGAAAGGAAAGGGUGGGCUUUCACCGAUCCCAAAAUCGCAUAUAUUGUAUUGUGUGCAAUGUUGCAAAAGGAUAACAGCGAA ..(((..(((..((((.(((((((((((((...(((.((....(((...(((((....)))))..))).....)).))).))))))...))))))))))))))....(.....).))).. ( -34.10) >DroSec_CAF1 17946 120 - 1 AAUGCAAGCAAUGCAUCUGCACACGCAAUAGAAAUGGGUGAAGGGAAAGGGUGGGCUUUCAGCGAUCCCAAAAUCGCAUAUAUUGUAUUGUGUGCAAUGUUGCAAAAGGAUAACAGCGAA ..(((..(((..((((.(((((((((((((...((((((...((((...(.(((....))).)..))))...))).))).))))))...))))))))))))))....(.....).))).. ( -32.10) >DroSim_CAF1 3096 120 - 1 AAUGCAAGCAAUGCAUCUGUACACGCAAUAGAAAUGGGAGAAGGGAAAGGGUGGGCUUUCAGCGAUCCCAAAAUCGCAUAUAUUGUAUUGUGUGCAAUGUUGCAAAAGGAUAACAGCGAA ..((((.(((((((((((((.......)))))..(((((.....(((((......))))).....))))).............)))))))).)))).(((((...........))))).. ( -31.20) >consensus AAUGCAAGCAAUGCAUCUGCACACGCAAUAGAAAUGGGAGAAGGGAAAGGGUGGGCUUUCAGCGAUCCCAAAAUCGCAUAUAUUGUAUUGUGUGCAAUGUUGCAAAAGGAUAACAGCGAA ..(((..(((..((((.(((((((((((((((..(((((.....(((((......))))).....)))))...)).....))))))...))))))))))))))....(.....).))).. (-31.05 = -31.17 + 0.11)

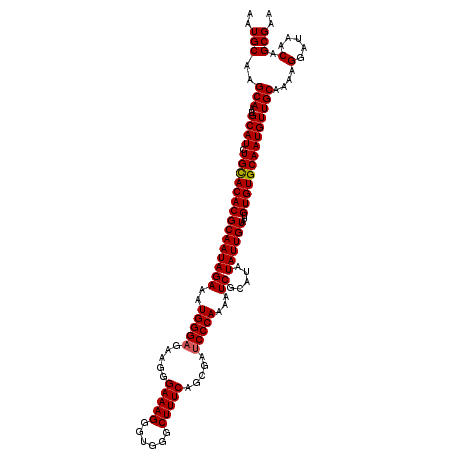
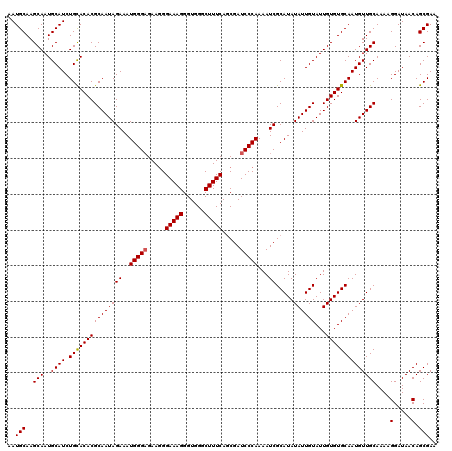
| Location | 6,995,630 – 6,995,750 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6995630 120 + 22224390 UAUGCGAUUUUGGGAUCGGUGAAAGCCCACCCUUUCCUUUCUCCCAUUUCUAUUGCGUGUGCAGAUGCAUUGCUUGCAUUCUAAUUUCCAUCAUUUUCGCAAAUAAUUGAUUUGUGUUCG ((((((((..(((((..((.(((((......)))))))...))))).....))))))))....((((((.....)))))).................(((((((.....))))))).... ( -28.00) >DroSec_CAF1 17986 120 + 1 UAUGCGAUUUUGGGAUCGCUGAAAGCCCACCCUUUCCCUUCACCCAUUUCUAUUGCGUGUGCAGAUGCAUUGCUUGCAUUCUAAUUUCCAUCAUUUUCGCAAAUAAUUGAUUUGUGUUCG ((((((((..((((......(((((......)))))......)))).....))))))))....((((((.....)))))).................(((((((.....))))))).... ( -25.00) >DroSim_CAF1 3136 120 + 1 UAUGCGAUUUUGGGAUCGCUGAAAGCCCACCCUUUCCCUUCUCCCAUUUCUAUUGCGUGUACAGAUGCAUUGCUUGCAUUCUAAUUUCCAUCAUUUUCGCAAAUAAUUGAUUUGUGUUCG ((((((((..(((((.....(((((......))))).....))))).....))))))))....((((((.....)))))).................(((((((.....))))))).... ( -26.90) >consensus UAUGCGAUUUUGGGAUCGCUGAAAGCCCACCCUUUCCCUUCUCCCAUUUCUAUUGCGUGUGCAGAUGCAUUGCUUGCAUUCUAAUUUCCAUCAUUUUCGCAAAUAAUUGAUUUGUGUUCG ((((((((..(((((.....(((((......))))).....))))).....))))))))....((((((.....)))))).................(((((((.....))))))).... (-25.70 = -26.03 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:03 2006