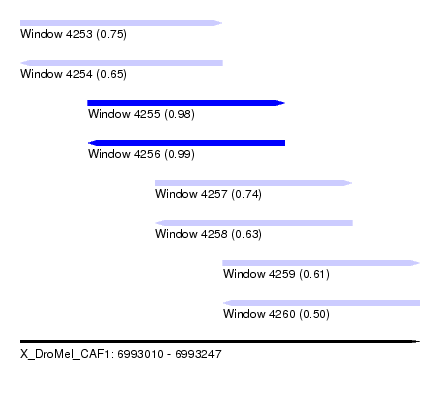
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,993,010 – 6,993,247 |
| Length | 237 |
| Max. P | 0.989101 |

| Location | 6,993,010 – 6,993,130 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -28.45 |
| Energy contribution | -28.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993010 120 + 22224390 UGAUUUGCUCAGUCAGGGUGGGAAAAAAUACACACCCAUAUAUAUGUAUGUAUGUAUAUUAUAUAUGUACUUACUCUUGGGUAGUUGGGGUAACCCGGCUUAUAAGCGUGAUAAUGAUGA ...........(((((((((............)))))........(((((((((((...)))))))))))((((.((((...(((((((....)))))))..)))).))))...)))).. ( -34.30) >DroSec_CAF1 15414 116 + 1 UGAUUUGCUCAGUCAGGGUGGGAAAAAAUACACACCCAUCUAUAUGUAUGCAUG----UUAUAUAUGUACAUACUUUUGGGUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGA ...((..(((.....)))..))........((((((((....(((((..(((((----(...)))))))))))....)))))(((((((....))))))).......))).......... ( -32.20) >DroSim_CAF1 518 116 + 1 UGAUUUGCUCAGUCAGGGUGGGAAAAAAUACACACCAAUAUAUAUGUAUGCAUG----UUAUAUAUGUACAUACUUUUGGGUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGA .....(((((((..((((((............))))..((((((((((......----.))))))))))....)).)))))))((((((....))))))..................... ( -28.50) >consensus UGAUUUGCUCAGUCAGGGUGGGAAAAAAUACACACCCAUAUAUAUGUAUGCAUG____UUAUAUAUGUACAUACUUUUGGGUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGA ...........(((((((((............)))))...((((((((((.........))))))))))..(((.((((...(((((((....)))))))..)))).)))....)))).. (-28.45 = -28.57 + 0.11)

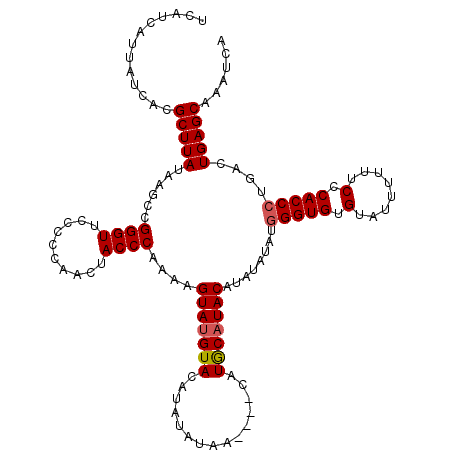
| Location | 6,993,010 – 6,993,130 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -19.68 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993010 120 - 22224390 UCAUCAUUAUCACGCUUAUAAGCCGGGUUACCCCAACUACCCAAGAGUAAGUACAUAUAUAAUAUACAUACAUACAUAUAUAUGGGUGUGUAUUUUUUCCCACCCUGACUGAGCAAAUCA .............(((((..((..((((..........))))..(((.((((((((((...(((((.((......)).)))))..)))))))))).))).....))...)))))...... ( -22.70) >DroSec_CAF1 15414 116 - 1 UCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUACCCAAAAGUAUGUACAUAUAUAA----CAUGCAUACAUAUAGAUGGGUGUGUAUUUUUUCCCACCCUGACUGAGCAAAUCA .............(((((......((((..........))))....(((((((.........----..)))))))........(((((.(........).)))))....)))))...... ( -23.20) >DroSim_CAF1 518 116 - 1 UCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUACCCAAAAGUAUGUACAUAUAUAA----CAUGCAUACAUAUAUAUUGGUGUGUAUUUUUUCCCACCCUGACUGAGCAAAUCA .............(((((....(.((((..................((((((.........)----)))))((((((((......))))))))........)))).)..)))))...... ( -21.40) >consensus UCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUACCCAAAAGUAUGUACAUAUAUAA____CAUGCAUACAUAUAUAUGGGUGUGUAUUUUUUCCCACCCUGACUGAGCAAAUCA .............(((((......((((..........))))....(((((((...............)))))))........(((((.(........).)))))....)))))...... (-19.68 = -20.13 + 0.45)

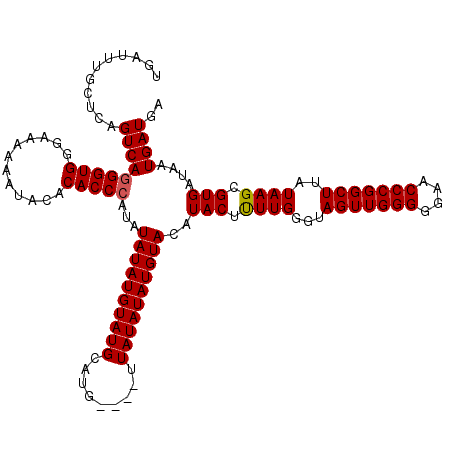
| Location | 6,993,050 – 6,993,167 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -36.25 |
| Energy contribution | -36.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993050 117 + 22224390 UAUAUGUAUGUAUGUAUAUUAUAUAUGUACUUACUCUUGGGUAGUUGGGGUAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUGUUUCCCAGCUU---GUCAGCUG .....(((((((((((...)))))))))))((((.((((...(((((((....)))))))..)))).))))..........(((((((..(((((......)))))...---))))))). ( -42.10) >DroSec_CAF1 15454 116 + 1 UAUAUGUAUGCAUG----UUAUAUAUGUACAUACUUUUGGGUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUG ((((((((((....----.))))))))))..(((.((((...(((((((....)))))))..)))).)))...........(((((((..(((((......)))))......))))))). ( -36.20) >DroSim_CAF1 558 116 + 1 UAUAUGUAUGCAUG----UUAUAUAUGUACAUACUUUUGGGUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUG ((((((((((....----.))))))))))..(((.((((...(((((((....)))))))..)))).)))...........(((((((..(((((......)))))......))))))). ( -36.20) >consensus UAUAUGUAUGCAUG____UUAUAUAUGUACAUACUUUUGGGUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUG ((((((((((.........))))))))))..(((.((((...(((((((....)))))))..)))).)))...........(((((((..(((((......)))))......))))))). (-36.25 = -36.03 + -0.22)



| Location | 6,993,050 – 6,993,167 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -27.91 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993050 117 - 22224390 CAGCUGAC---AAGCUGGGAAACAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUACCCCAACUACCCAAGAGUAAGUACAUAUAUAAUAUACAUACAUACAUAUA ..((((((---...(((((......)))))..))))))...........((..(((....))).((((..........))))..))(((.(((..((((...))))..))).)))..... ( -29.50) >DroSec_CAF1 15454 116 - 1 CAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUACCCAAAAGUAUGUACAUAUAUAA----CAUGCAUACAUAUA ..((((((......(((((......)))))..))))))...............(((....))).((((..........))))....(((((((.........----..)))))))..... ( -29.80) >DroSim_CAF1 558 116 - 1 CAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUACCCAAAAGUAUGUACAUAUAUAA----CAUGCAUACAUAUA ..((((((......(((((......)))))..))))))...............(((....))).((((..........))))....(((((((.........----..)))))))..... ( -29.80) >consensus CAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUACCCAAAAGUAUGUACAUAUAUAA____CAUGCAUACAUAUA ..((((((......(((((......)))))..))))))...............(((....))).((((..........))))....(((((((...............)))))))..... (-27.91 = -28.03 + 0.11)


| Location | 6,993,090 – 6,993,207 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -41.90 |
| Energy contribution | -41.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993090 117 + 22224390 GUAGUUGGGGUAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUGUUUCCCAGCUU---GUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUU ..(((((((....)))))))..(((((((......((((..(((((((..(((((......)))))...---)))))))((((.....))))..))))((....)).))).))))..... ( -42.90) >DroSec_CAF1 15490 120 + 1 GUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUU ..(((((((....)))))))..(((((((......((((..(((((((..(((((......)))))......)))))))((((.....))))..))))((....)).))).))))..... ( -42.40) >DroSim_CAF1 594 120 + 1 GUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUU ..(((((((....)))))))..(((((((......((((..(((((((..(((((......)))))......)))))))((((.....))))..))))((....)).))).))))..... ( -42.40) >consensus GUAGUUGGGGGAACCCGGCUUAUAAGCGUGAUAAUGAUGAUGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUU ..(((((((....)))))))..(((((((......((((..(((((((..(((((......)))))......)))))))((((.....))))..))))((....)).))).))))..... (-41.90 = -41.90 + -0.00)

| Location | 6,993,090 – 6,993,207 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -33.13 |
| Energy contribution | -33.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993090 117 - 22224390 AAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGAC---AAGCUGGGAAACAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUACCCCAACUAC ...((((((..((((.((......)).))))...))))))..((((((---...(((((......)))))..))))))...............(((....))).(((....)))...... ( -35.50) >DroSec_CAF1 15490 120 - 1 AAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUAC ..((((.(((.(((.....)))..(((((((...........)))))))....)))(((...((((((.((((....)).))...........(((....))).)))))).))))))).. ( -34.80) >DroSim_CAF1 594 120 - 1 AAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUAC ..((((.(((.(((.....)))..(((((((...........)))))))....)))(((...((((((.((((....)).))...........(((....))).)))))).))))))).. ( -34.80) >consensus AAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCAUCAUCAUUAUCACGCUUAUAAGCCGGGUUCCCCCAACUAC ...((((((..((((.((......)).))))...))))))..((((((......(((((......)))))..))))))...............(((....))).(((....)))...... (-33.13 = -33.13 + -0.00)



| Location | 6,993,130 – 6,993,247 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -46.23 |
| Consensus MFE | -42.17 |
| Energy contribution | -42.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993130 117 + 22224390 UGGCUGACCUCUGGGCUGUUUCCCAGCUU---GUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUUUGACCAUUGGUCAGGAGCAAGACGGGAGCAGCACGUGUCC .(((((((..(((((......)))))...---)))))))((((.....))))((((((((....((..((.(((..(((((((((...))))))))).))).))...)).))).))))). ( -45.30) >DroSec_CAF1 15530 119 + 1 UGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUUUGACCAUUGGUCAGGAGCAAGA-GGGAGCAGCACGUGUCC ..((((.(((((..((((((..((((((.......)))((((((((((.((((.((......)).)))).)......))))))))).)))..)))))).)))-))...))))........ ( -46.70) >DroSim_CAF1 634 119 + 1 UGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUUUGACCAUUGGUCAGGAGCAAGA-GGGAGCAGCACGUGUCC ..((((.(((((..((((((..((((((.......)))((((((((((.((((.((......)).)))).)......))))))))).)))..)))))).)))-))...))))........ ( -46.70) >consensus UGGCUGACCUCUGGGCUCUUUCCCAGCUUGUUGUCAGCUGGUCAAAAUGGCCGACAUUGCCUUGGCGGCGACUUAACUUUUGACCAUUGGUCAGGAGCAAGA_GGGAGCAGCACGUGUCC .(((((((..(((((......)))))......)))))))((((.....))))((((((((....)))((..((...(((((((((...)))))))))......))..)).....))))). (-42.17 = -42.17 + -0.00)

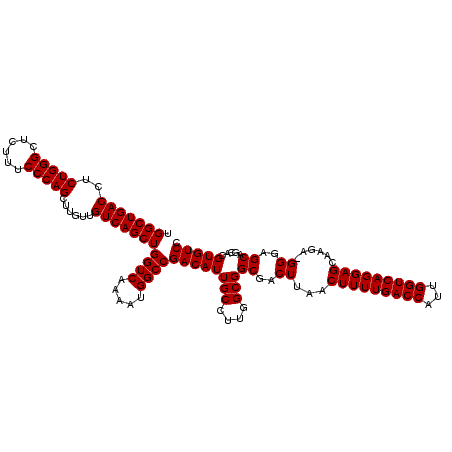
| Location | 6,993,130 – 6,993,247 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -37.83 |
| Energy contribution | -37.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6993130 117 - 22224390 GGACACGUGCUGCUCCCGUCUUGCUCCUGACCAAUGGUCAAAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGAC---AAGCUGGGAAACAGCCCAGAGGUCAGCCA .((((..((((((...((.((((((..(((((...)))))..)).)))).))..))...)))).))))((.(.....).)).((((((---...(((((......)))))..)))))).. ( -41.20) >DroSec_CAF1 15530 119 - 1 GGACACGUGCUGCUCCC-UCUUGCUCCUGACCAAUGGUCAAAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCA ......(.((((...((-(((.((((.(..(((.((((((((((......)((((.((......)).))))..)))))))))(((.......))))))..).))))..))))).))))). ( -40.80) >DroSim_CAF1 634 119 - 1 GGACACGUGCUGCUCCC-UCUUGCUCCUGACCAAUGGUCAAAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCA ......(.((((...((-(((.((((.(..(((.((((((((((......)((((.((......)).))))..)))))))))(((.......))))))..).))))..))))).))))). ( -40.80) >consensus GGACACGUGCUGCUCCC_UCUUGCUCCUGACCAAUGGUCAAAAGUUAAGUCGCCGCCAAGGCAAUGUCGGCCAUUUUGACCAGCUGACAACAAGCUGGGAAAGAGCCCAGAGGUCAGCCA ((.((.(.((............)).).)).))..((((((((((......)((((.((......)).))))..)))))))))((((((......(((((......)))))..)))))).. (-37.83 = -37.83 + 0.00)

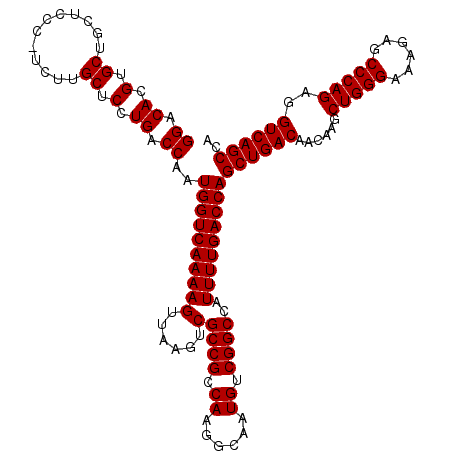
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:53 2006