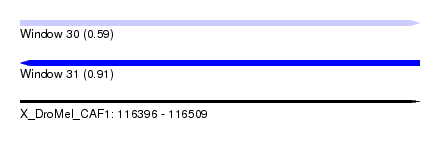
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 116,396 – 116,509 |
| Length | 113 |
| Max. P | 0.913124 |

| Location | 116,396 – 116,509 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -22.67 |
| Energy contribution | -24.67 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
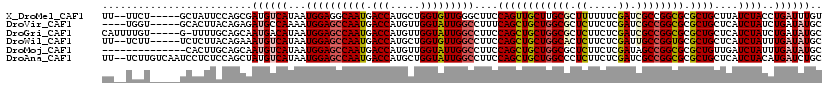
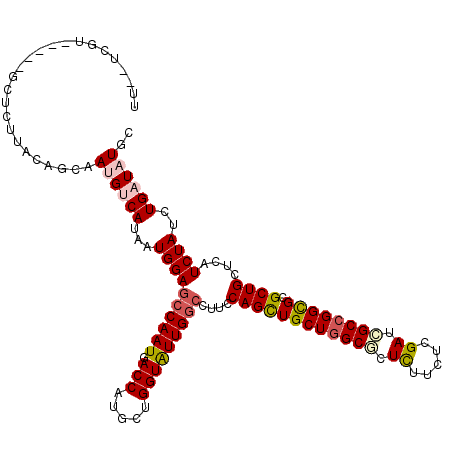
>X_DroMel_CAF1 116396 113 + 22224390 ACAAAUCAGGUAGAUAAGCAGCGCGCCGGCGAUCGAAAAAAGCGCAAGCAACUGGAAGCCCAACACCAGCAUGGUCAUUGCCUCCAUUAUGACAUCGCUGGAAUAGC-----AGAA--AA ....................((...((((((((.(......((....))...((((.((.....(((.....)))....)).))))......)))))))))....))-----....--.. ( -29.50) >DroVir_CAF1 5151 111 + 1 GCAUAUCAGAUAGAUGAGCAGCGCGCCGGCGAUCGAGAAGAGCGCCAGCAGCUGAAAGGCCAAUACCAACAUGGUCAUUGGCUCCAUUUUGGCAUCUCUGUAAGUGC-----ACCA---- ((((..((((..((((..((((..((.((((.((.....)).)))).)).))))...((((((((((.....))).))))))).........))))))))...))))-----....---- ( -42.20) >DroGri_CAF1 5647 114 + 1 GCAUAUCAGAUAGAUGAGCAGCGCGCCGGCGAUCGAGAAGAGCGCCAGCAGCUGGAAGGCCAAUACCAACAUGGUCAUUGGCUCCAUUAUGUCAUUGCUGCAAAA-C-----ACAAAAUG ...((((.....)))).(((((((((.((((.((.....)).)))).)).))((((..(((((((((.....))).))))))))))..........)))))....-.-----........ ( -39.20) >DroWil_CAF1 5660 113 + 1 GCAUAUCAAAUAGAUGAGCAGCGCACCGGCAAUCGAGAAGAGUGCCAGCAGCUGGAAGGCCAACACCAGCAUGGUCAUUGGCUCCAUUAUGACAUUUCUGUAAGAGA-----AAGA--AA ((.((((.....)))).))(((((...((((.((.....)).)))).)).)))(((..(((((.(((.....)))..)))))))).........(((((.....)))-----))..--.. ( -29.60) >DroMoj_CAF1 5357 106 + 1 GCAUAUCAAAUAGAUCAACAGCGCGCCGGCUAUCGAGAAGAGCGCCAGCAGCUGGAAGGCCAAUACCAACAUGGUCAUUGGCUCCAUUAUGACAUUGCUGCAAGUG-------------- (((..(((..........((((..((.(((..((.....))..))).)).))))...((((((((((.....))).)))))))......)))...)))........-------------- ( -31.90) >DroAna_CAF1 5142 118 + 1 GCAGAUCAUGUAGAUGAGCAGCGCGCCGGCGAUCGAGAAGAGGGCCAGCAGCUGGAAGGCCAAUACCAGCAUGGUCAUUGGCUCCAUUAUGACAUAGCUGGAGAGGAUUGACAAGA--AA .(((...((((.((((..((((..((.(((..((.....))..))).)).))))...((((((((((.....))).))))))).))))...))))..)))................--.. ( -36.20) >consensus GCAUAUCAGAUAGAUGAGCAGCGCGCCGGCGAUCGAGAAGAGCGCCAGCAGCUGGAAGGCCAAUACCAACAUGGUCAUUGGCUCCAUUAUGACAUUGCUGCAAGAGC_____AAAA__AA ..................((((((((.((((.((.....)).)))).)).))((((..(((((((((.....))).))))))))))..........)))).................... (-22.67 = -24.67 + 2.00)

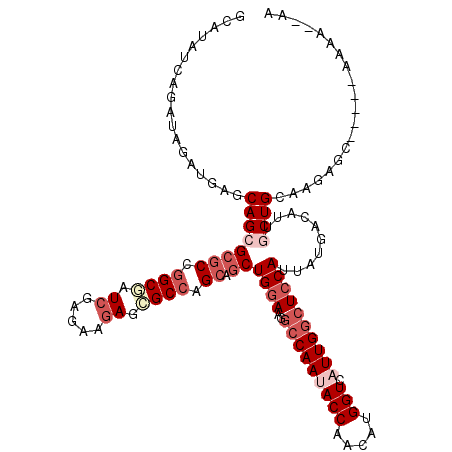
| Location | 116,396 – 116,509 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -31.44 |
| Energy contribution | -32.13 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 116396 113 - 22224390 UU--UUCU-----GCUAUUCCAGCGAUGUCAUAAUGGAGGCAAUGACCAUGCUGGUGUUGGGCUUCCAGUUGCUUGCGCUUUUUUCGAUCGCCGGCGCGCUGCUUAUCUACCUGAUUUGU ..--....-----.......(((((.........(((((((....(((.....))).....)))))))...(((.(((.((.....)).))).))).)))))(..(((.....)))..). ( -28.10) >DroVir_CAF1 5151 111 - 1 ----UGGU-----GCACUUACAGAGAUGCCAAAAUGGAGCCAAUGACCAUGUUGGUAUUGGCCUUUCAGCUGCUGGCGCUCUUCUCGAUCGCCGGCGCGCUGCUCAUCUAUCUGAUAUGC ----....-----(((..(.((((((((((.....)).(((((((.((.....)))))))))....((((((((((((.((.....)).)))))))).))))..))))..)))))..))) ( -39.80) >DroGri_CAF1 5647 114 - 1 CAUUUUGU-----G-UUUUGCAGCAAUGACAUAAUGGAGCCAAUGACCAUGUUGGUAUUGGCCUUCCAGCUGCUGGCGCUCUUCUCGAUCGCCGGCGCGCUGCUCAUCUAUCUGAUAUGC ........-----.-....(((((..........(((((((((((.((.....)))))))))..))))((.(((((((.((.....)).))))))))))))))..(((.....))).... ( -42.10) >DroWil_CAF1 5660 113 - 1 UU--UCUU-----UCUCUUACAGAAAUGUCAUAAUGGAGCCAAUGACCAUGCUGGUGUUGGCCUUCCAGCUGCUGGCACUCUUCUCGAUUGCCGGUGCGCUGCUCAUCUAUUUGAUAUGC ..--...(-----(((.....))))((((((.((((((((((((.(((.....)))))))))....((((((((((((.((.....)).)))))))).))))....)))))))))))).. ( -38.10) >DroMoj_CAF1 5357 106 - 1 --------------CACUUGCAGCAAUGUCAUAAUGGAGCCAAUGACCAUGUUGGUAUUGGCCUUCCAGCUGCUGGCGCUCUUCUCGAUAGCCGGCGCGCUGUUGAUCUAUUUGAUAUGC --------------........(((.(((((.(((((((((((((.((.....)))))))))....(((((((((((..((.....))..))))))).))))....)))))))))))))) ( -39.80) >DroAna_CAF1 5142 118 - 1 UU--UCUUGUCAAUCCUCUCCAGCUAUGUCAUAAUGGAGCCAAUGACCAUGCUGGUAUUGGCCUUCCAGCUGCUGGCCCUCUUCUCGAUCGCCGGCGCGCUGCUCAUCUACAUGAUCUGC ..--....((((((.....(((((...(((((..((....)))))))...))))).))))))....(((((((((((..((.....))..))))))).)))).((((....))))..... ( -36.10) >consensus UU__UCGU_____GCUCUUACAGCAAUGUCAUAAUGGAGCCAAUGACCAUGCUGGUAUUGGCCUUCCAGCUGCUGGCGCUCUUCUCGAUCGCCGGCGCGCUGCUCAUCUAUCUGAUAUGC .........................((((((...((((((((((.(((.....)))))))))....((((((((((((.((.....)).)))))))).))))....))))..)))))).. (-31.44 = -32.13 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:40 2006