
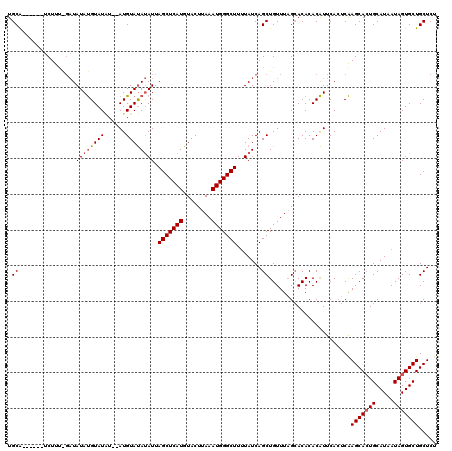
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,940,523 – 6,940,672 |
| Length | 149 |
| Max. P | 0.972579 |

| Location | 6,940,523 – 6,940,632 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.23 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6940523 109 - 22224390 UGCA------UCUUC---UAUAUGUAUAU--AUGUAUACAUUAGCUCAUGUACUUAAAUGGGCUUUUAUCAGCUGUUUAGAACACACAUUCACUCAAGCACUGCAUAAUAGUGCUGCUCU .((.------..(((---((.(((((((.--...))))))).(((((((........))))))).............)))))..............(((((((.....)))))))))... ( -23.90) >DroSec_CAF1 25524 112 - 1 UGCG------UCUUUAGAUAUAUGUAUGU--AUGUAUAUAUUAGCUCAUGUACUUAAAUGGGCUUUUAUCAGCUGUUUAGCACACACAUUCACUCAAGCACUGCAUAAUAGUGCUGCUCU .(((------((....)))...((.((((--.(((..(((..(((((((........)))))))..)))..(((....)))))).)))).))....(((((((.....)))))))))... ( -26.30) >DroEre_CAF1 34110 120 - 1 UGCAUACAUAUCUUUGGAUAUACGUAUCUAGAUGUACAUAUUAGCUCAUGUACUUAAAUGGGCUUAUAUCAGCUGUUUCGCACACACGUUCAAGCGAGCAGUGCAUAAUAGUGCUGCUCU .......((((((..(((((....)))))))))))..((((.(((((((........))))))).))))..(((....((......))....)))((((((..(......)..)))))). ( -35.20) >consensus UGCA______UCUUU_GAUAUAUGUAUAU__AUGUAUAUAUUAGCUCAUGUACUUAAAUGGGCUUUUAUCAGCUGUUUAGCACACACAUUCACUCAAGCACUGCAUAAUAGUGCUGCUCU .((................((((((((......)))))))).(((((((........))))))).......)).......................(((((((.....)))))))..... (-17.69 = -18.47 + 0.78)


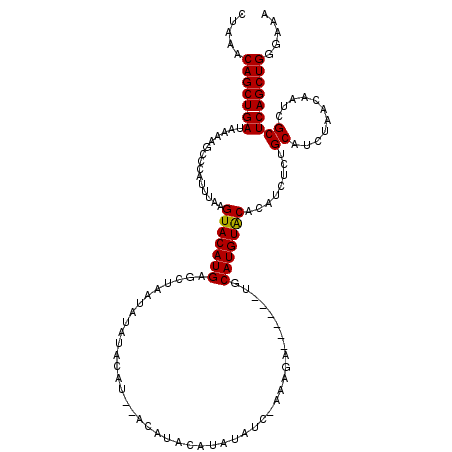
| Location | 6,940,563 – 6,940,672 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6940563 109 + 22224390 CUAAACAGCUGAUAAAAGCCCAUUUAAGUACAUGAGCUAAUGUAUACAU--AUAUACAUAUA---GAAGA------UGCAUGUACACAUCUGUGCAUCUAACAAUCGCUCAGCUGGGAAA .....(((((((....(((.(((........))).))).(((((((...--.)))))))...---..(((------((((((........))))))))).........)))))))..... ( -28.20) >DroSec_CAF1 25564 112 + 1 CUAAACAGCUGAUAAAAGCCCAUUUAAGUACAUGAGCUAAUAUAUACAU--ACAUACAUAUAUCUAAAGA------CGCAUGUACACAUCUCUGCAUCUAACAAUCGCUCAGCUGGGAAA .....(((((((...............(((((((..((.((((((....--......))))))....)).------..)))))))........((...........)))))))))..... ( -21.00) >DroEre_CAF1 34150 120 + 1 CGAAACAGCUGAUAUAAGCCCAUUUAAGUACAUGAGCUAAUAUGUACAUCUAGAUACGUAUAUCCAAAGAUAUGUAUGCAUGUGCACAUCUCUGCGUCUAACAGUCGCUCAGCUGGGUAA .....((((((((((.(((.(((........))).))).)))(((((((....((((((((........))))))))..))))))).......(((.(.....).))))))))))..... ( -30.70) >consensus CUAAACAGCUGAUAAAAGCCCAUUUAAGUACAUGAGCUAAUAUAUACAU__ACAUACAUAUAUC_AAAGA______UGCAUGUACACAUCUCUGCAUCUAACAAUCGCUCAGCUGGGAAA .....(((((((...............(((((((............................................)))))))........((...........)))))))))..... (-17.59 = -17.37 + -0.22)

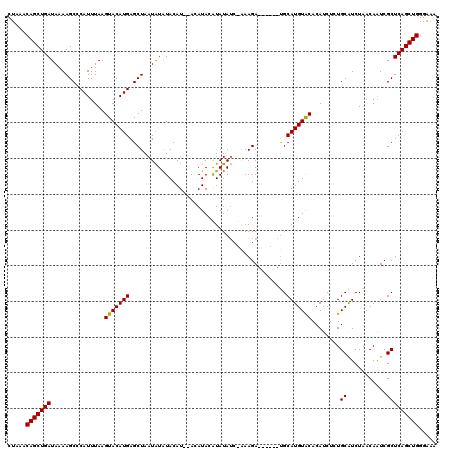
| Location | 6,940,563 – 6,940,672 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -27.19 |
| Energy contribution | -26.64 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6940563 109 - 22224390 UUUCCCAGCUGAGCGAUUGUUAGAUGCACAGAUGUGUACAUGCA------UCUUC---UAUAUGUAUAU--AUGUAUACAUUAGCUCAUGUACUUAAAUGGGCUUUUAUCAGCUGUUUAG .....(((((((((.(((....)))))...((((((((((((.(------((...---.....).)).)--)))))))))))(((((((........)))))))....)))))))..... ( -31.50) >DroSec_CAF1 25564 112 - 1 UUUCCCAGCUGAGCGAUUGUUAGAUGCAGAGAUGUGUACAUGCG------UCUUUAGAUAUAUGUAUGU--AUGUAUAUAUUAGCUCAUGUACUUAAAUGGGCUUUUAUCAGCUGUUUAG .....(((((((((.(((....)))))...((((((((((((((------(...((....))...))))--)))))))))))(((((((........)))))))....)))))))..... ( -34.60) >DroEre_CAF1 34150 120 - 1 UUACCCAGCUGAGCGACUGUUAGACGCAGAGAUGUGCACAUGCAUACAUAUCUUUGGAUAUACGUAUCUAGAUGUACAUAUUAGCUCAUGUACUUAAAUGGGCUUAUAUCAGCUGUUUCG .....((((((((((.(.....).)))....(((((((..((.(((((((((....)))))..)))).))..)))))))...(((((((........)))))))....)))))))..... ( -34.60) >consensus UUUCCCAGCUGAGCGAUUGUUAGAUGCAGAGAUGUGUACAUGCA______UCUUU_GAUAUAUGUAUAU__AUGUAUAUAUUAGCUCAUGUACUUAAAUGGGCUUUUAUCAGCUGUUUAG .....((((((((((.(.....).)))...(((((((((((..............................)))))))))))(((((((........)))))))....)))))))..... (-27.19 = -26.64 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:39 2006