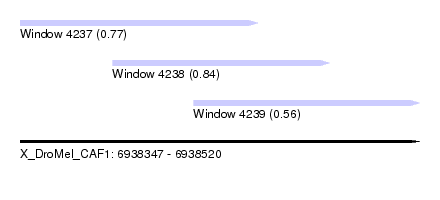
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,938,347 – 6,938,520 |
| Length | 173 |
| Max. P | 0.839707 |

| Location | 6,938,347 – 6,938,450 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Mean single sequence MFE | -37.71 |
| Consensus MFE | -32.58 |
| Energy contribution | -31.89 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6938347 103 + 22224390 AGGCAGAUGUUCCGCAUUCUCGGAAGCUGGAUGCCGGCAAGGUGAUAGUUCUAUUUU-----UUUGGGCCGGGUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAG------------ ..((..(((((((((...(((((..(((((...)))))...........((((....-----..)))))))))..((((.....)))).....))))))))).))...------------ ( -35.40) >DroSec_CAF1 23399 102 + 1 AGGCAGAUGUUCCGCAUUCUCGGAAGCUGGAUGCCGGCAAGGUGAUAGUUUUAUUU------UUUGGGCCGGGUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAG------------ ..((..(((((((((...(((((..(((((...)))))(((((((.....))))))------).....)))))..((((.....)))).....))))))))).))...------------ ( -36.00) >DroEre_CAF1 31962 106 + 1 AGGCAGAUGUUCCGCAUUCUCGGAAGCUGGAUGCCGGCAAGGUGAUAGCCCUAUUU------UUUGGGCCG-UUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAGCUGC-------A ..((((.(((..(((((((.((...(((((...))))).........((((.....------...))))))-..))))).....((((....))))...))..)))..))))-------. ( -38.70) >DroYak_CAF1 24579 119 + 1 AGGCAGAUGUUCCGCAUUCUCGGAAGCUGGAUGCCGGCAAGGUGAUAGCCCUACUUUUUAUUUUUGGGCCG-UUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAGCUGCAAACUGCA ..((((..(((((((.(((......(((((...))))).....((..((((..............))))..-)).((((.....)))))))..)))))))(..(....)..)...)))). ( -40.74) >consensus AGGCAGAUGUUCCGCAUUCUCGGAAGCUGGAUGCCGGCAAGGUGAUAGCCCUAUUU______UUUGGGCCG_GUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAG____________ ..((..(((((((((.(((.((...(((((...))))).........((((..............))))))....((((.....)))))))..))))))))).))............... (-32.58 = -31.89 + -0.69)

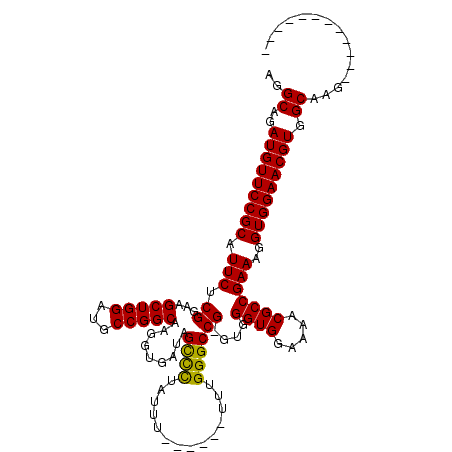
| Location | 6,938,387 – 6,938,481 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -21.08 |
| Energy contribution | -20.64 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6938387 94 + 22224390 GGUGAUAGUUCUAUUUU-----UUUGGGCCGGGUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAG---------------------CUGCAUGUUAAUUACUUGGAAGCAAGCUGCA .......((((....((-----(((.(.((....)).).)))))((((....))))))))(..((..(---------------------((.((.((.....)).))..)))..))..). ( -27.80) >DroSec_CAF1 23439 93 + 1 GGUGAUAGUUUUAUUU------UUUGGGCCGGGUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAG---------------------CUGCAAGUUAAUUACUUGGAAGCAAGCUGCA .......(((((((((------((((((((.....)))(.....).))))))))))))))(..((..(---------------------((.(((((.....)))))..)))..))..). ( -30.80) >DroEre_CAF1 32002 106 + 1 GGUGAUAGCCCUAUUU------UUUGGGCCG-UUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAGCUGC-------AACCGGCAAGCUGCAAGUUAAUUACUUGGAAGCAAGCUGCA ...((..((((.....------...))))..-))..........((((....))))....(..((..((((.-------...))))..(((.(((((.....)))))..)))..))..). ( -36.50) >DroYak_CAF1 24619 119 + 1 GGUGAUAGCCCUACUUUUUAUUUUUGGGCCG-UUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAGCUGCAAACUGCAAGCGGCAAGCUGCAAGUUAAUUACUUGGAAGCAAGCUGCA (((..((((.((((.((((((((((.(((.(-((........)))))).)))))))))).))))...))))...)))....(((((..(((.(((((.....)))))..)))..))))). ( -40.40) >consensus GGUGAUAGCCCUAUUU______UUUGGGCCG_GUGGGUGGAAAACGCCGAAAGGUGGAACGUGGCAAG_____________________CUGCAAGUUAAUUACUUGGAAGCAAGCUGCA .......((((..............)))).......(..(....((((....)))).(((...(((.(((.................))))))..))).....((((....)))))..). (-21.08 = -20.64 + -0.44)

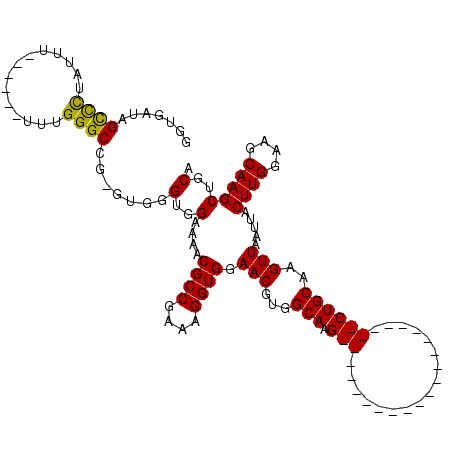

| Location | 6,938,422 – 6,938,520 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6938422 98 + 22224390 AAAACGCCGAAAGGUGGAACGUGGCAAG---------------------CUGCAUGUUAAUUACUUGGAAGCAAGCUGCAUUUUGUAUUGCUACCCGAUUUAAAUUCUG-UGAACAUCUU ....((((....))))....((((((((---------------------(...((((......((((....))))..))))...)).))))))).............((-....)).... ( -22.60) >DroSec_CAF1 23473 98 + 1 AAAACGCCGAAAGGUGGAACGUGGCAAG---------------------CUGCAAGUUAAUUACUUGGAAGCAAGCUGCAUUUUGUAUUGCUACCCGACUUAAAUUCUG-UGAACAUGUU ....((((....)))).(((((((((((---------------------((.(((((.....)))))..)))....(((.....)))))))))).............((-....)).))) ( -25.80) >DroEre_CAF1 32035 113 + 1 AAAACGCCGAAAGGUGGAACGUGGCAAGCUGC-------AACCGGCAAGCUGCAAGUUAAUUACUUGGAAGCAAGCUGCAUUUUGUAUCGCUACCCGACUUAAUUUCCGUUGAAUAUGUU ..((((((....)))((((.(..((..((((.-------...))))..(((.(((((.....)))))..)))..))..)...(((..(((.....)))..))).)))))))......... ( -30.80) >DroYak_CAF1 24658 111 + 1 AAAACGCCGAAAGGUGGAACGUGGCAAGCUGCAAACUGCAAGCGGCAAGCUGCAAGUUAAUUACUUGGAAGCAAGCUGCAUUUUGUAUCGCUACCCGACUUAA---------AAAUUCUU ....((((....))))....(((((....(((.....))).(((((..(((.(((((.....)))))..)))..)))))..........))))).........---------........ ( -35.20) >consensus AAAACGCCGAAAGGUGGAACGUGGCAAG_____________________CUGCAAGUUAAUUACUUGGAAGCAAGCUGCAUUUUGUAUCGCUACCCGACUUAAAUUCUG_UGAACAUCUU ....((((....))))....(((((...........................(((((.....)))))((.(((((......))))).))))))).......................... (-21.10 = -21.10 + -0.00)

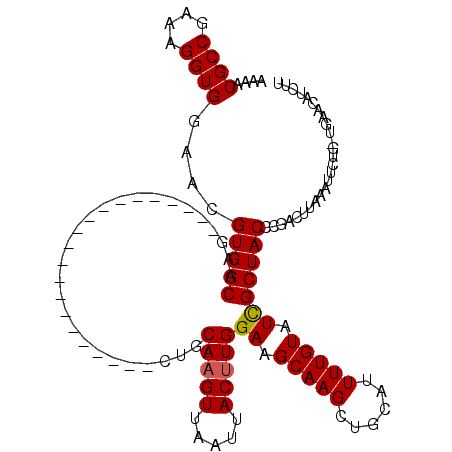
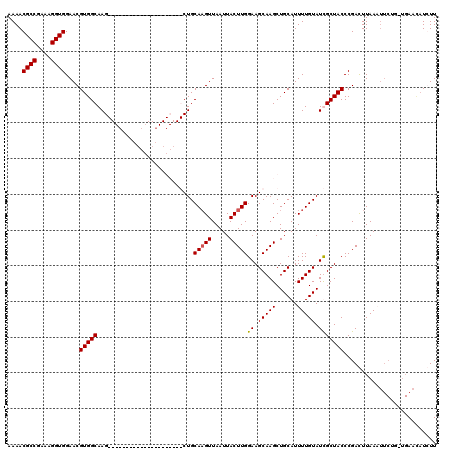
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:35 2006