
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,931,356 – 6,931,456 |
| Length | 100 |
| Max. P | 0.992614 |

| Location | 6,931,356 – 6,931,456 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6931356 100 + 22224390 GGUUGUUAUAAUAAU---------UCUCGAGUUCUCGAUAUCGAUAUAGAC----------ACAAUACAUAUAUAUCGAUCGUUGCUUUUUG-GCGGCAGCAACAACAACACACAACUUG .((((((........---------..((((....)))).(((((((((...----------..........))))))))).((((((....)-)))))))))))................ ( -22.82) >DroSec_CAF1 16446 94 + 1 GGUUGUUAU---AAU---------UCUCGAGUUCUGGAGAUCGAUAUAGAC----------ACA-UAC--AUAUAUCGACCGUUGCUUUUUG-CCGGCAGCAACAACAACACACAACUUG .((((((..---...---------((((.(....).))))((((((((...----------...-...--.))))))))..((((((.....-..))))))))))))............. ( -23.10) >DroSim_CAF1 15042 94 + 1 GGUUGUUAU---AAU---------UCUCGAGUUCUGGAGAUCGAUAUAGAC----------ACA-UAC--AUAUAUCGAUCGUUGCUUUUUG-CCGGCAGCAACAACAACACACAACUUG .((((((..---...---------(((.(....).)))((((((((((...----------...-...--.))))))))))((((((.....-..))))))))))))............. ( -24.80) >DroEre_CAF1 24786 102 + 1 GGUUGUUAU---AUU---------UCUCGAGUUCUCGAGAUCGAUAUAGAACACAUAAUGUACA-UAC--AUAUAUCGAUUGUUGCUUUCGGGUCAGCAACA---ACAACACACAACUUG ((((((...---...---------((((((....))))))((((((((....((.....))...-...--.))))))))((((((((........)))))))---)......)))))).. ( -26.50) >DroYak_CAF1 17147 101 + 1 GGUUGUUUU---AUUUUGACGAGCUCUCAAGUUCUCGAGAUCGAUAUAUAU--------------UGC--AUAUAUCGAUCGUUGCUUUCGGGUCAGCAACAACAACAACACACAACUUG .((((((..---...((((.(((((....)))))))))(((((((((((..--------------...--)))))))))))((((((........))))))))))))............. ( -31.40) >consensus GGUUGUUAU___AAU_________UCUCGAGUUCUCGAGAUCGAUAUAGAC__________ACA_UAC__AUAUAUCGAUCGUUGCUUUUUG_CCGGCAGCAACAACAACACACAACUUG .((((((...............................((((((((((.......................))))))))))((((((........))))))...)))))).......... (-21.32 = -21.32 + 0.00)

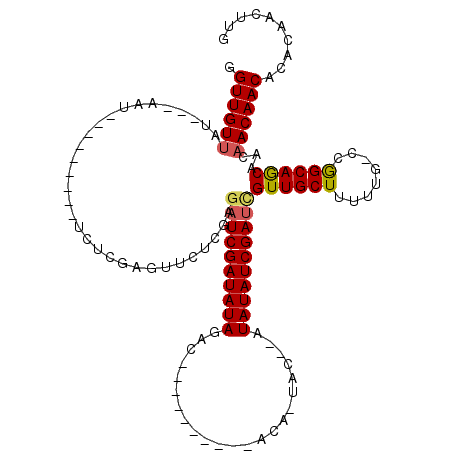
| Location | 6,931,356 – 6,931,456 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6931356 100 - 22224390 CAAGUUGUGUGUUGUUGUUGCUGCCGC-CAAAAAGCAACGAUCGAUAUAUAUGUAUUGU----------GUCUAUAUCGAUAUCGAGAACUCGAGA---------AUUAUUAUAACAACC (((..(((((((((((((((((.....-.....)))))))).)))))))))....)))(----------((.((((..(((.((((....))))..---------)))..)))))))... ( -24.20) >DroSec_CAF1 16446 94 - 1 CAAGUUGUGUGUUGUUGUUGCUGCCGG-CAAAAAGCAACGGUCGAUAUAU--GUA-UGU----------GUCUAUAUCGAUCUCCAGAACUCGAGA---------AUU---AUAACAACC ...(((((..((((((.((((.....)-)))..))))))((((((((((.--(..-...----------..)))))))))))..............---------...---...))))). ( -24.00) >DroSim_CAF1 15042 94 - 1 CAAGUUGUGUGUUGUUGUUGCUGCCGG-CAAAAAGCAACGAUCGAUAUAU--GUA-UGU----------GUCUAUAUCGAUCUCCAGAACUCGAGA---------AUU---AUAACAACC ...(((((..((((((.((((.....)-)))..))))))((((((((((.--(..-...----------..)))))))))))..............---------...---...))))). ( -24.60) >DroEre_CAF1 24786 102 - 1 CAAGUUGUGUGUUGU---UGUUGCUGACCCGAAAGCAACAAUCGAUAUAU--GUA-UGUACAUUAUGUGUUCUAUAUCGAUCUCGAGAACUCGAGA---------AAU---AUAACAACC ...((((((((((.(---(((((((........))))))))((((((((.--.((-((.....))))..)...)))))))((((((....))))))---------)))---)).))))). ( -31.10) >DroYak_CAF1 17147 101 - 1 CAAGUUGUGUGUUGUUGUUGUUGCUGACCCGAAAGCAACGAUCGAUAUAU--GCA--------------AUAUAUAUCGAUCUCGAGAACUUGAGAGCUCGUCAAAAU---AAAACAACC ...(((((...(((((...((((((........))))))(((((((((((--(..--------------.)))))))))))).((((..(....)..))))....)))---)).))))). ( -30.90) >consensus CAAGUUGUGUGUUGUUGUUGCUGCCGA_CAAAAAGCAACGAUCGAUAUAU__GUA_UGU__________GUCUAUAUCGAUCUCGAGAACUCGAGA_________AUU___AUAACAACC ...(((((((((((((((((((...........)))))))).))))))................................((((((....))))))..................))))). (-18.88 = -19.60 + 0.72)

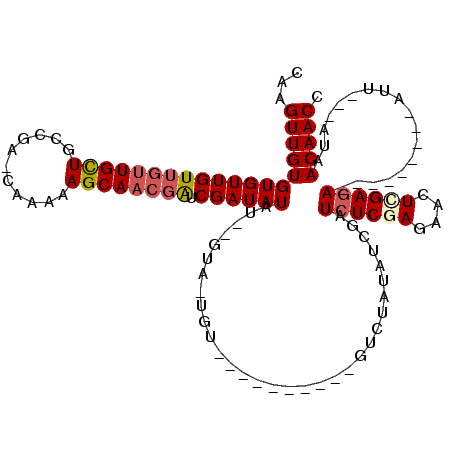

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:26 2006