
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,931,011 – 6,931,122 |
| Length | 111 |
| Max. P | 0.988580 |

| Location | 6,931,011 – 6,931,122 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -45.08 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
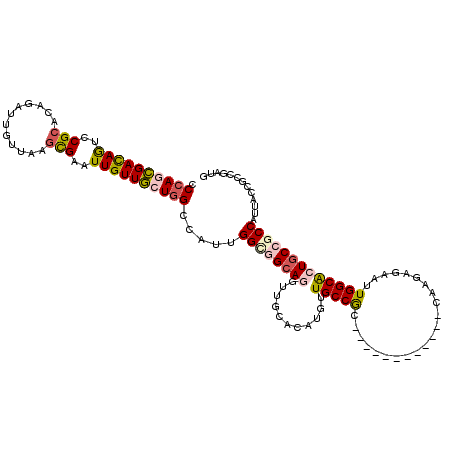
>X_DroMel_CAF1 6931011 111 + 22224390 CCCAGCGACAAUCCGCAGAGAUUAUUCAGCGAGUUGUUGCUGGCCAUCGGCGGCAGGUGGCACAUGUUGCCGCC---------CCCUAAAUUGUUGGCACUCCCACCAUUUCCGCCGCUG .((((((((((..(((.(((....))).)))..))))))))))....(((((((.(((((((.....)))))))---------........((.(((.....))).)).....))))))) ( -48.90) >DroVir_CAF1 22261 102 + 1 CCAAUCGAUAGGCCGCACAGAUUGUUCAGCGAAUUGUUGCUGGCCAUUGGCGGCAUAUUGCAGAUGUUGCCGC------------CAAGGGAAUUGGCGCUGCCGCCAUUACCG------ ..........((((((((((.(((.....))).))).))).)))).(((((((((((((...)))).))))))------------)))((....(((((....)))))...)).------ ( -40.90) >DroPse_CAF1 15874 108 + 1 CCCAGCGACAGUCCGCAUAGAUUGCUAAGCGAAUUGUUGCUGGUCAUUGGCAGCAGGUUGCACAUGUUGCCGC------------CAAGAUUAUUGGCACUGCCGCCAUUGGCGCCGAUG .(((((((((((.(((.(((....))).))).))))))))))).(((((((.((((((.((....)).)))((------------(((.....)))))..))).(((...)))))))))) ( -50.80) >DroWil_CAF1 19874 106 + 1 CCCAAUGAUAGACCGCACAGGUUGUUCAAUGAAUUGUUACUGGCCAUCGGUGGCAUAUUGCACAUAUUGCCACC---------GCCAAGGGAAUUGGCACUGCCGCCAUUACCGC----- ..((.(((..((((.....))))..))).)).........((((....(((((((............)))))))---------(((((.....)))))......)))).......----- ( -32.80) >DroAna_CAF1 14867 120 + 1 CCCAGCGACAGGCCGCACAGAUUGUUAAGCGAAUUGUUGCUGGUCAUGGGCGGCAGGUUGCACAUGUUGCCGCCACCGCCGCCGCCAAGGGAAUUGGCACUGCCACCAUUACCACCGAUG .((((((((((..(((............)))..))))))))))..(((((((((.(((.(((.....))).)))...))))))(((((.....)))))........)))........... ( -46.30) >DroPer_CAF1 15885 108 + 1 CCCAGCGACAGUCCGCAUAGAUUGCUAAGCGAAUUGUUGCUGGUCAUUGGCAGCAGGUUGCACAUGUUGCCGC------------CAAGAUUAUUGGCACUGCCGCCAUUGGCGCCGAUG .(((((((((((.(((.(((....))).))).))))))))))).(((((((.((((((.((....)).)))((------------(((.....)))))..))).(((...)))))))))) ( -50.80) >consensus CCCAGCGACAGUCCGCACAGAUUGUUAAGCGAAUUGUUGCUGGCCAUUGGCGGCAGGUUGCACAUGUUGCCGC____________CAAGAGAAUUGGCACUGCCGCCAUUACCGCCGAUG .((((((((((..(((............)))..)))))))))).....((((((((...........(((((......................)))))))))))))............. (-27.18 = -27.88 + 0.70)


| Location | 6,931,011 – 6,931,122 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -26.26 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6931011 111 - 22224390 CAGCGGCGGAAAUGGUGGGAGUGCCAACAAUUUAGGG---------GGCGGCAACAUGUGCCACCUGCCGCCGAUGGCCAGCAACAACUCGCUGAAUAAUCUCUGCGGAUUGUCGCUGGG ((.((((((....(((((.(..(((..(......)..---------)))(....)...).)))))..)))))).)).(((((.((((..(((.((......)).)))..)))).))))). ( -51.40) >DroVir_CAF1 22261 102 - 1 ------CGGUAAUGGCGGCAGCGCCAAUUCCCUUG------------GCGGCAACAUCUGCAAUAUGCCGCCAAUGGCCAGCAACAAUUCGCUGAACAAUCUGUGCGGCCUAUCGAUUGG ------(((((.((((((((.((((((.....)))------------)))(((.....)))....))))))))..((((.(((...(((........)))...))))))))))))..... ( -40.00) >DroPse_CAF1 15874 108 - 1 CAUCGGCGCCAAUGGCGGCAGUGCCAAUAAUCUUG------------GCGGCAACAUGUGCAACCUGCUGCCAAUGACCAGCAACAAUUCGCUUAGCAAUCUAUGCGGACUGUCGCUGGG .........((.(((((((((((((((.....)))------------)))(((.....)))...))))))))).)).(((((.(((.(((((.(((....))).))))).))).))))). ( -46.20) >DroWil_CAF1 19874 106 - 1 -----GCGGUAAUGGCGGCAGUGCCAAUUCCCUUGGC---------GGUGGCAAUAUGUGCAAUAUGCCACCGAUGGCCAGUAACAAUUCAUUGAACAACCUGUGCGGUCUAUCAUUGGG -----...((..((((.((...(((((.....)))))---------(((((((.(((.....))))))))))).).))))...))...(((.(((...(((.....)))...))).))). ( -32.50) >DroAna_CAF1 14867 120 - 1 CAUCGGUGGUAAUGGUGGCAGUGCCAAUUCCCUUGGCGGCGGCGGUGGCGGCAACAUGUGCAACCUGCCGCCCAUGACCAGCAACAAUUCGCUUAACAAUCUGUGCGGCCUGUCGCUGGG (((.(((((((..(((.(((.((((((.....))))))((.((....)).))......))).)))))))))).))).(((((.(((..((((............))))..))).))))). ( -48.10) >DroPer_CAF1 15885 108 - 1 CAUCGGCGCCAAUGGCGGCAGUGCCAAUAAUCUUG------------GCGGCAACAUGUGCAACCUGCUGCCAAUGACCAGCAACAAUUCGCUUAGCAAUCUAUGCGGACUGUCGCUGGG .........((.(((((((((((((((.....)))------------)))(((.....)))...))))))))).)).(((((.(((.(((((.(((....))).))))).))).))))). ( -46.20) >consensus CAUCGGCGGUAAUGGCGGCAGUGCCAAUAACCUUG____________GCGGCAACAUGUGCAACCUGCCGCCAAUGACCAGCAACAAUUCGCUGAACAAUCUGUGCGGACUGUCGCUGGG ............((((((((((((.......................)))(((.....)))...)))))))))....(((((.(((..((((.((......)).))))..))).))))). (-26.26 = -26.68 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:24 2006