
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,926,761 – 6,926,861 |
| Length | 100 |
| Max. P | 0.564815 |

| Location | 6,926,761 – 6,926,861 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.83 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -8.78 |
| Energy contribution | -8.37 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6926761 100 - 22224390 ----CUUG-AAAAGUAUAAUACGAUAUCCUUGCAA--------UACAAUGGUCAUAGUAA-----AAAGGUGUUAACGAUUACCCUGAUCAAUAGCAAGAGUAUCUCAGAACUUCGGA- ----.(((-(...(((...)))((((..(((((.(--------(....((((((..((((-----...(.......)..))))..)))))))).)))))..))))))))..(....).- ( -19.00) >DroPse_CAF1 11104 111 - 1 AGUUCUUUUGAACCCA-GAAUCGAUGUCCUUGCAGGGUGCAGAAGCAGAACACACCAAAAAAAAAAAAGAUG------AUCCAAUCAGACAACUGCAAGGGCAUCG-AGGGCUUCGGAC ......(((((((((.-...(((((((((((((((((((.............))))............(((.------.....)))......))))))))))))))-)))).)))))). ( -40.22) >DroSec_CAF1 11780 100 - 1 ----CUUG-AAAAGUAUAAUACGAUAUCUUUGCAA--------UACAAUGGUCAUAGUGA-----AAAUGUGUUAACGAUUCCCCUGAUCAAUGGCAAGAGUAUCUUUGAACUUCGGA- ----.(((-(..(((.(((...(((((..((((..--------..((.((((((..(.((-----(..(((....))).))))..)))))).))))))..))))).))).))))))).- ( -17.40) >DroEre_CAF1 20226 100 - 1 ----CUUGUGAAAGUGUAAAACGAUAUCCUUGCAA--------UACAAUGCUCAUUGUGA-----AAAGAUGUUAAC-AUUCCCCUGAUCAAUGGCAAGAAUAUCCUCGAACUUCGGA- ----....((((.((.......(((((.(((((..--------((((((....)))))).-----...((((....)-))).............))))).))))).....))))))..- ( -17.00) >DroYak_CAF1 12425 102 - 1 ----CUUGUGAAAGUAUAAUACGAUAUCCUUGCAA--------UACAAUGGUCAUCGUGAG----AAAUAUGUAAACGAUUCCCCUGAUCAAUGGCAAGAAUAUCUUCGAGCUUCGGA- ----..................(((((.(((((..--------..((.((((((((....)----)....((....)).......)))))).))))))).)))))((((.....))))- ( -18.90) >DroPer_CAF1 11122 111 - 1 AGUCCCUUUGAACCCA-GAAUCGAUAUCCUUGCAGGGAGCAGAAGCAGUCCACACCAAAAAAAAAAAAGAUG------AUCCAAUCAGACAACUGCAAGGGCAUCG-AGGGCUUCGGAC .((((....((((((.-...(((((..(((((((((((((....))..))).................(((.------.....)))......))))))))..))))-)))).))))))) ( -35.60) >consensus ____CUUGUGAAAGUAUAAAACGAUAUCCUUGCAA________UACAAUGCUCAUCGUAA_____AAAGAUGUUAAC_AUUCCCCUGAUCAAUGGCAAGAGUAUCUUAGAACUUCGGA_ ......................(((((.(((((.............................................................))))).)))))......(....).. ( -8.78 = -8.37 + -0.42)

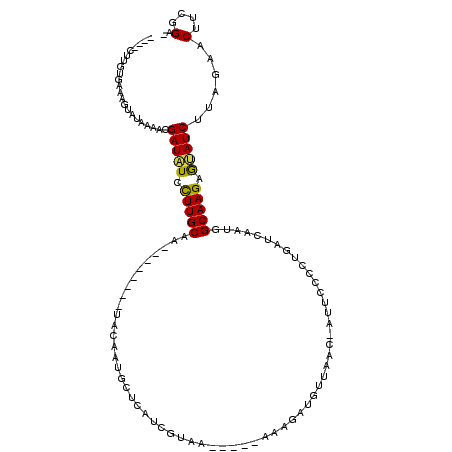
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:21 2006