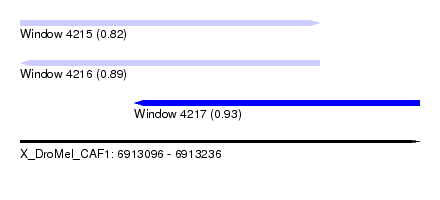
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,913,096 – 6,913,236 |
| Length | 140 |
| Max. P | 0.933748 |

| Location | 6,913,096 – 6,913,201 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -16.90 |
| Energy contribution | -18.15 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6913096 105 + 22224390 GAAAUUUGCCUACAGCCACCAUAAAUCAGAUUCAAUUUAAAUUAUCUGGCCACUGGUUGCCGGCCAUAAAUUACU-UAUCGUUUCAUUUGACCAGACAAGG--------------UUUUU (((((..(((..((((((.......((((((............))))))....))))))..))).((((.....)-))).)))))....((((......))--------------))... ( -18.10) >DroSec_CAF1 37454 105 + 1 GAAAUUUGCCUACAGCCACCAUAAAUCAGAUUCAAUUUAAAUUAUCUGGACACUGGUUGCCGGCCAUAAAUAACU-UAUCGUUUCACUUGGCCAGACAACG--------------UUUUU ((((((((.((...((.((((....((((((............))))))....)))).)).(((((.....(((.-....))).....))))))).))).)--------------)))). ( -22.50) >DroSim_CAF1 21604 105 + 1 GAAAUUUGCCUACAGCCACCAUAAAUCAGAUUCAAUUUAAAUUAUCUGGACACUGGUUGCCGGCCAUAAAUAACU-UAUCGUUUCACUUGGCCAGACAACG--------------UUUUU ((((((((.((...((.((((....((((((............))))))....)))).)).(((((.....(((.-....))).....))))))).))).)--------------)))). ( -22.50) >DroYak_CAF1 41389 120 + 1 GAAAUUUGCCUACGGCCAUCAUAAAUCAGAUUUAAUUUAAAUUAUCUGCUCACUGCCUGCCGGCCAUAAAUAACCAAAUCGUUUCACUUGGCCAGGCAACAGUCAACAGGCAACAGUUUU .....((((((...............(((((............)))))...((((..(((((((((.....(((......))).....))))).)))).))))....))))))....... ( -29.60) >consensus GAAAUUUGCCUACAGCCACCAUAAAUCAGAUUCAAUUUAAAUUAUCUGGACACUGGUUGCCGGCCAUAAAUAACU_UAUCGUUUCACUUGGCCAGACAACG______________UUUUU .....(((.((...((.((((....((((((............))))))....)))).)).(((((.....(((......))).....))))))).)))..................... (-16.90 = -18.15 + 1.25)


| Location | 6,913,096 – 6,913,201 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.22 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6913096 105 - 22224390 AAAAA--------------CCUUGUCUGGUCAAAUGAAACGAUA-AGUAAUUUAUGGCCGGCAACCAGUGGCCAGAUAAUUUAAAUUGAAUCUGAUUUAUGGUGGCUGUAGGCAAAUUUC .....--------------.((((((...((....))...))))-)).........((((....)((((.((((...((((............))))..)))).))))..)))....... ( -23.20) >DroSec_CAF1 37454 105 - 1 AAAAA--------------CGUUGUCUGGCCAAGUGAAACGAUA-AGUUAUUUAUGGCCGGCAACCAGUGUCCAGAUAAUUUAAAUUGAAUCUGAUUUAUGGUGGCUGUAGGCAAAUUUC .....--------------..(((((((((((.....(((....-.))).....)))))(((.((((..(((.((((............)))))))...)))).)))..))))))..... ( -27.10) >DroSim_CAF1 21604 105 - 1 AAAAA--------------CGUUGUCUGGCCAAGUGAAACGAUA-AGUUAUUUAUGGCCGGCAACCAGUGUCCAGAUAAUUUAAAUUGAAUCUGAUUUAUGGUGGCUGUAGGCAAAUUUC .....--------------..(((((((((((.....(((....-.))).....)))))(((.((((..(((.((((............)))))))...)))).)))..))))))..... ( -27.10) >DroYak_CAF1 41389 120 - 1 AAAACUGUUGCCUGUUGACUGUUGCCUGGCCAAGUGAAACGAUUUGGUUAUUUAUGGCCGGCAGGCAGUGAGCAGAUAAUUUAAAUUAAAUCUGAUUUAUGAUGGCCGUAGGCAAAUUUC ....(((..(((((((.(((((((((.(((((.....(((......))).....))))))))).))))).)))).......(((((((....)))))))....)))..)))......... ( -35.80) >consensus AAAAA______________CGUUGUCUGGCCAAGUGAAACGAUA_AGUUAUUUAUGGCCGGCAACCAGUGUCCAGAUAAUUUAAAUUGAAUCUGAUUUAUGGUGGCUGUAGGCAAAUUUC .....................(((((((((((.....(((......))).....)))))(((.((((.....(((((............))))).....)))).)))..))))))..... (-26.48 = -26.22 + -0.25)


| Location | 6,913,136 – 6,913,236 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6913136 100 - 22224390 UAACAAAAGCCCUGGGACUACAAAAACAC-----AUAAAAAAAAA--------------CCUUGUCUGGUCAAAUGAAACGAUA-AGUAAUUUAUGGCCGGCAACCAGUGGCCAGAUAAU ........((((((((....)........-----...........--------------..(((.(((((((......((....-.))......)))))))))))))).)))........ ( -19.40) >DroSec_CAF1 37494 94 - 1 AGGCAAAGGCCCUGGGACUACAAA-ACAC----------AAAAAA--------------CGUUGUCUGGCCAAGUGAAACGAUA-AGUUAUUUAUGGCCGGCAACCAGUGUCCAGAUAAU .(((....)))((((((((.....-....----------......--------------.((((.(((((((.....(((....-.))).....))))))))))).))).)))))..... ( -25.13) >DroSim_CAF1 21644 94 - 1 AGGCAAAGGCCCUGGGACUACAAA-ACGC----------AAAAAA--------------CGUUGUCUGGCCAAGUGAAACGAUA-AGUUAUUUAUGGCCGGCAACCAGUGUCCAGAUAAU .(((....)))((((((((.....-..(.----------......--------------)((((.(((((((.....(((....-.))).....))))))))))).))).)))))..... ( -26.10) >DroYak_CAF1 41429 119 - 1 AGCCAAAGGCCCUGGGUUUUGGGA-ACACAAACAAUAAAAAAAACUGUUGCCUGUUGACUGUUGCCUGGCCAAGUGAAACGAUUUGGUUAUUUAUGGCCGGCAGGCAGUGAGCAGAUAAU ..((((((.(....).))))))..-.......(((((........))))).(((((.(((((((((.(((((.....(((......))).....))))))))).))))).)))))..... ( -38.50) >consensus AGGCAAAGGCCCUGGGACUACAAA_ACAC__________AAAAAA______________CGUUGUCUGGCCAAGUGAAACGAUA_AGUUAUUUAUGGCCGGCAACCAGUGUCCAGAUAAU ...........(((((..........).................................((((((.(((((.....(((......))).....)))))))))))......))))..... (-15.09 = -15.53 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:15 2006