
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,912,837 – 6,913,027 |
| Length | 190 |
| Max. P | 0.893216 |

| Location | 6,912,837 – 6,912,947 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.75 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6912837 110 + 22224390 CAGACAAACUCACGUUUUCGUUCUUACUGUUUUAUCAGCCGUCUGGCAUGGCAUUUACAUAU----------AAUCGAACGAUUCAGUGUGUUUGGGUUUUUUUUUUAUAUAUAUUGUUU ....(((((.(((....((((((....((......))(((((.....)))))..........----------....))))))....))).)))))......................... ( -23.00) >DroSec_CAF1 37200 105 + 1 CAGACAAACUCACGUUUUCGUUCUUACUGUUUUAUCAGCCGUCUGGCAAGGCAUUUACAUAU----------AAUCGAACGAUUCAGUGUGUUUGUGCUUUUUUU-----UAUAUGGUUU ...((((((.(((....((((((...(((......)))..((((....))))..........----------....))))))....))).)))))).........-----.......... ( -24.60) >DroSim_CAF1 21350 105 + 1 CAGACAAACUCACGUUUUCGUUCUUACUGUUUUAUCAGCAGUCUGGCAAGGCAUUUACAUAU----------AAUCGAACGAUUCAGUGUGUUUGGGUUUUUUUU-----UAUAUUGUUU ....(((((.(((....((((((...(((......)))..((((....))))..........----------....))))))....))).)))))..........-----.......... ( -23.50) >DroYak_CAF1 41112 117 + 1 CAGGCAAUCUCACGUUUUCGUUGUUACUGUUUUAUCAGCCGUCUGGCAAGGCAUUUACAUAUACAUAUAUACAAUCGAACGAUUUAGUGUGUUUGCGUCUUGUUUUCGU---UAUUUUUG ...((((.(.(((....(((((((...((((((..(((....)))..))))))...))((((....)))).......)))))....))).).)))).............---........ ( -20.80) >consensus CAGACAAACUCACGUUUUCGUUCUUACUGUUUUAUCAGCCGUCUGGCAAGGCAUUUACAUAU__________AAUCGAACGAUUCAGUGUGUUUGGGUUUUUUUU_____UAUAUUGUUU ...((((((.(((....((((((...(((......)))..((((....))))........................))))))....))).))))))........................ (-19.62 = -20.75 + 1.12)

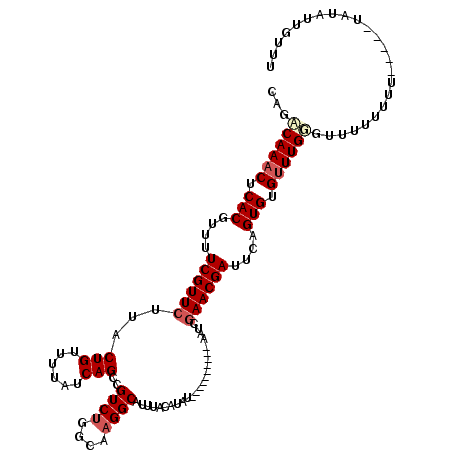
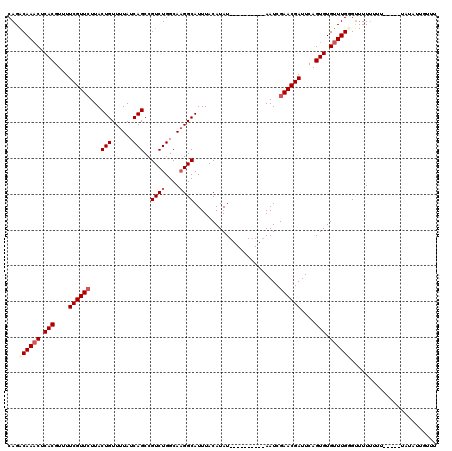
| Location | 6,912,837 – 6,912,947 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6912837 110 - 22224390 AAACAAUAUAUAUAAAAAAAAAACCCAAACACACUGAAUCGUUCGAUU----------AUAUGUAAAUGCCAUGCCAGACGGCUGAUAAAACAGUAAGAACGAAAACGUGAGUUUGUCUG .........................(((((.(((....((((((....----------...(((...((.((.(((....))))).))..)))....))))))....))).))))).... ( -19.10) >DroSec_CAF1 37200 105 - 1 AAACCAUAUA-----AAAAAAAGCACAAACACACUGAAUCGUUCGAUU----------AUAUGUAAAUGCCUUGCCAGACGGCUGAUAAAACAGUAAGAACGAAAACGUGAGUUUGUCUG ..........-----......((.((((((.(((....((((((....----------....((((.....))))......((((......))))..))))))....))).)))))))). ( -21.10) >DroSim_CAF1 21350 105 - 1 AAACAAUAUA-----AAAAAAAACCCAAACACACUGAAUCGUUCGAUU----------AUAUGUAAAUGCCUUGCCAGACUGCUGAUAAAACAGUAAGAACGAAAACGUGAGUUUGUCUG ..........-----..........(((((.(((....((((((....----------....((((.....)))).....(((((......))))).))))))....))).))))).... ( -19.40) >DroYak_CAF1 41112 117 - 1 CAAAAAUA---ACGAAAACAAGACGCAAACACACUAAAUCGUUCGAUUGUAUAUAUGUAUAUGUAAAUGCCUUGCCAGACGGCUGAUAAAACAGUAACAACGAAAACGUGAGAUUGCCUG ........---.............((((.(.(((....(((((..(((((......((((......))))...(((....))).......)))))...)))))....))).).))))... ( -19.10) >consensus AAACAAUAUA_____AAAAAAAACCCAAACACACUGAAUCGUUCGAUU__________AUAUGUAAAUGCCUUGCCAGACGGCUGAUAAAACAGUAAGAACGAAAACGUGAGUUUGUCUG ........................((((((.(((....((((((..................((((.....))))......((((......))))..))))))....))).))))))... (-15.60 = -16.73 + 1.12)

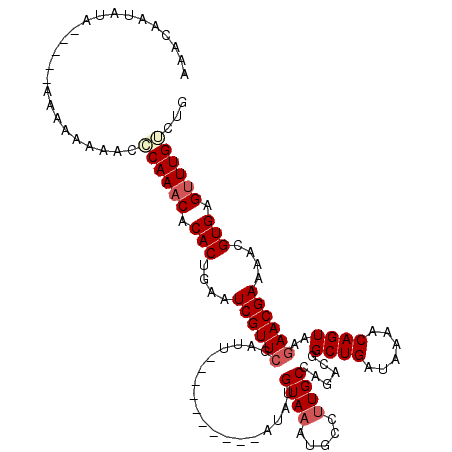
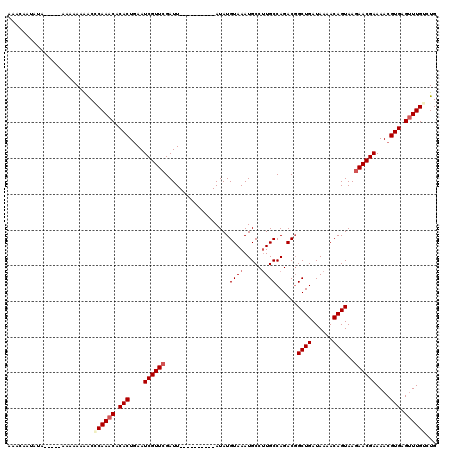
| Location | 6,912,877 – 6,912,987 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6912877 110 + 22224390 GUCUGGCAUGGCAUUUACAUAU----------AAUCGAACGAUUCAGUGUGUUUGGGUUUUUUUUUUAUAUAUAUUGUUUUAGAUUCUUUUUUCGUUUUGGCUUUUGCCGAGACACGGGA ((((((((.(((....((((((----------((..(((.(((((((.....))))))).)))..))))))...........((........))))....)))..)))).))))...... ( -24.40) >DroSec_CAF1 37240 105 + 1 GUCUGGCAAGGCAUUUACAUAU----------AAUCGAACGAUUCAGUGUGUUUGUGCUUUUUUU-----UAUAUGGUUUUUGAUUAUUUUUUCGUUUUGGCAUUUGCCGAGACACCGGA .(((((.(((((((..((((((----------((((....))))..))))))..)))))))....-----........................((((((((....)))))))).))))) ( -31.30) >DroSim_CAF1 21390 105 + 1 GUCUGGCAAGGCAUUUACAUAU----------AAUCGAACGAUUCAGUGUGUUUGGGUUUUUUUU-----UAUAUUGUUUUUGAUUUUUUUUUCGUUUUGGCAUUUGCCGAGACACCGGA .(((((.(((((....((((((----------((((....))))..))))))....)))))....-----........................((((((((....)))))))).))))) ( -25.20) >DroYak_CAF1 41152 117 + 1 GUCUGGCAAGGCAUUUACAUAUACAUAUAUACAAUCGAACGAUUUAGUGUGUUUGCGUCUUGUUUUCGU---UAUUUUUGUUUCUUUUUUUUUUAUUUCGACUUUUGCCGAGACAUCGGC (((.((((((((((......)).((.((((((((((....))))..)))))).)).))))))))...(.---.((....))..)...............)))....(((((....))))) ( -19.80) >consensus GUCUGGCAAGGCAUUUACAUAU__________AAUCGAACGAUUCAGUGUGUUUGGGUUUUUUUU_____UAUAUUGUUUUUGAUUUUUUUUUCGUUUUGGCAUUUGCCGAGACACCGGA .(((((.(((((....((((((..........((((....))))..))))))....))))).................................((((((((....)))))))).))))) (-18.27 = -18.77 + 0.50)



| Location | 6,912,907 – 6,913,027 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.71 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6912907 120 + 22224390 GAUUCAGUGUGUUUGGGUUUUUUUUUUAUAUAUAUUGUUUUAGAUUCUUUUUUCGUUUUGGCUUUUGCCGAGACACGGGAGAGUAAUUAAUACAGAAUUACAAGCUGAAACUGAUUGAUU ...(((((..((((.(((((.........................(((((..(.((((((((....)))))))))..)))))((((((.......))))))))))).))))..))))).. ( -28.40) >DroSec_CAF1 37270 115 + 1 GAUUCAGUGUGUUUGUGCUUUUUUU-----UAUAUGGUUUUUGAUUAUUUUUUCGUUUUGGCAUUUGCCGAGACACCGGAGAGUAAUUAAUACAGAAUUACAAGCUGAAACUGAUUGAUU ...(((((..(((((((.((((...-----......((..(((((((((((((.((((((((....))))))))...))))))))))))).)))))).)))))))....)))))...... ( -30.30) >DroSim_CAF1 21420 115 + 1 GAUUCAGUGUGUUUGGGUUUUUUUU-----UAUAUUGUUUUUGAUUUUUUUUUCGUUUUGGCAUUUGCCGAGACACCGGAGAGUAAUGCAAACAGAAUUACAAGCUGAAACUGAUUGAUG ...(((((..(((((...((((.((-----(.((((((((((((........))((((((((....))))))))....)))))))))).))).))))...)))))....)))))...... ( -27.40) >DroYak_CAF1 41192 117 + 1 GAUUUAGUGUGUUUGCGUCUUGUUUUCGU---UAUUUUUGUUUCUUUUUUUUUUAUUUCGACUUUUGCCGAGACAUCGGCGAGUAAUUGAUGCAGAAUUACAAGCUGAAACUGAUUGAUU ..((((((.((((((((((......(((.---.((...................))..)))..((((((((....)))))))).....)))))))....))).))))))........... ( -24.31) >consensus GAUUCAGUGUGUUUGGGUUUUUUUU_____UAUAUUGUUUUUGAUUUUUUUUUCGUUUUGGCAUUUGCCGAGACACCGGAGAGUAAUUAAUACAGAAUUACAAGCUGAAACUGAUUGAUU ...(((((..((((.(((((..................................((((((((....))))))))........((((((.......))))))))))).))))..))))).. (-19.86 = -20.80 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:13 2006