
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,892,715 – 6,892,954 |
| Length | 239 |
| Max. P | 0.988294 |

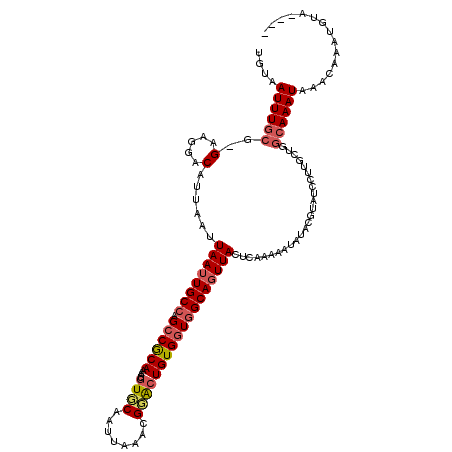
| Location | 6,892,715 – 6,892,834 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.68 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6892715 119 + 22224390 UGUAAUUUGCG-GGUGGACAUUAAUUAAUUGCCAGCCGCAAAUGGUUCAAUUAAACGGACUGUGGUGGCAGUUACUUAAAAAUAUACAUAUCCUUGCUGGCAAAUAAACAAAUGUACAUA (((.((((((.-((((((..((((.(((((((((.(((((....((((........)))))))))))))))))).))))...........)))..))).))))))..))).......... ( -30.92) >DroSec_CAF1 17047 116 + 1 UGUAAUUUGCG-GGGGGACAUUAAUUAAUUGCCAGCCGCAAAAUGUUCAAUUAAACGGGAUGUGAUGGCAGUUACUCAAAAAUAUACGUAUCCUUGCUGGCAAAUAAAGAAAUUUAC--- (((.....(((-(((..((......(((((((((..((((...((((......))))...)))).))))))))).............))..))))))..)))..((((....)))).--- ( -25.81) >DroEre_CAF1 16504 113 + 1 CGUAAUUUGCG-GAAGGACAUUAAUUAAAUGCCCGCCACAAAAUGUGCAAUUACACGGGCUGUGGUGGCAGUUACUCAAAAAUAUACGUAUCCUUGCUAGCAAAUAAACAAAUG------ ....(((((((-((((((...........((((.((((((...((((......))))...))))))))))..(((............)))))))).)).)))))).........------ ( -28.40) >DroYak_CAF1 19573 105 + 1 UGUAAUUUGCGCGAAGGACAUUAAUUAAUUGCACGCCGCAAAAUGUGCAAUUAAAUGCACUGUGGUGGCAGUUACUCAAAAAUAUAA--------------AAAUAAACAAAUGUA-AUG ....(((((.((....).)......(((((((.(((((((....(((((......))))))))))))))))))).............--------------.......)))))...-... ( -25.10) >consensus UGUAAUUUGCG_GAAGGACAUUAAUUAAUUGCCAGCCGCAAAAUGUGCAAUUAAACGGACUGUGGUGGCAGUUACUCAAAAAUAUACGUAUCCUUGCUGGCAAAUAAACAAAUGUA____ ....((((((..(.....)......((((((((.((((((....((((........)))))))))))))))))).........................))))))............... (-16.61 = -17.68 + 1.06)

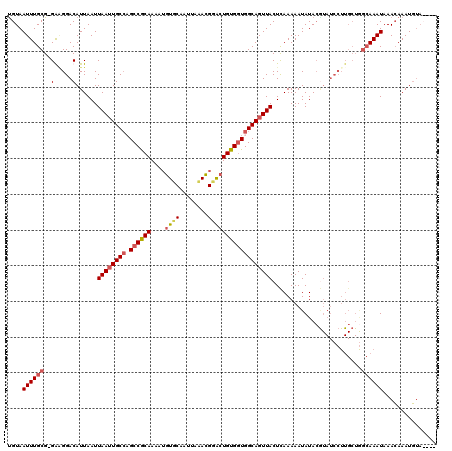
| Location | 6,892,834 – 6,892,954 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.03 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6892834 120 + 22224390 AACAUAUGUGCUGCAGUUGUAAUUUGUUUACCAUUUUUAUUUGAUACUUAACAAAUGACUGCAACUAAAUGAAAUUCUCUGUAACACCAGAGGAAUCUAAUGCUCCUCUGGAAUUCCGUU ..(((.((.(.(((((((...(((((((....(((.......)))....)))))))))))))).))).)))...............((((((((.........))))))))......... ( -26.10) >DroSec_CAF1 17163 117 + 1 ---AUAUGUGCUGCAGUUGUAAUUUGUUUCCUAUUUUUAUUUGACACCUAACAAAUGACUGCAACUAAAUGGAAUUCUCUGUAACCCCAGAGGAAUCGAAUGCUCCUCUGGAAUUGCGUU ---........(((((((...(((((((.....................)))))))))))))).................((((..((((((((.........))))))))..))))... ( -27.30) >DroEre_CAF1 16617 115 + 1 ----UACGUGCUGCAGUUGUAGUUUGUUUCCCAUU-UUAUUGGACACUUAACAAAUGACUGCAAAUAAAUGGAAUUUUCUGUAACCCCAGAGGAAUCGAAUGCUCCUCUGGAAUUCCGUU ----.......(((((((...(((((((..(((..-....)))......))))))))))))))....(((((((((..........((((((((.........))))))))))))))))) ( -33.10) >DroYak_CAF1 19678 120 + 1 UGUGUAUGUGUUGCAGUUGUAAUUCGUUUCCCAUUUUUAUUUGACACUUAACAAAUGACUGCAAAUAAAUUGCAUUUUCUGUAGCGCCAGAGGCAUCGAAUGCUCCUCUGGAAUUCUACU ...(((...(((((((.(((((((..(((..((...(((((((........))))))).)).)))..)))))))....))))))).((((((((((...))))..)))))).....))). ( -29.00) >consensus ___AUAUGUGCUGCAGUUGUAAUUUGUUUCCCAUUUUUAUUUGACACUUAACAAAUGACUGCAAAUAAAUGGAAUUCUCUGUAACCCCAGAGGAAUCGAAUGCUCCUCUGGAAUUCCGUU ...........(((((((...(((((((.....................)))))))))))))).......................((((((((.........))))))))......... (-22.54 = -22.85 + 0.31)


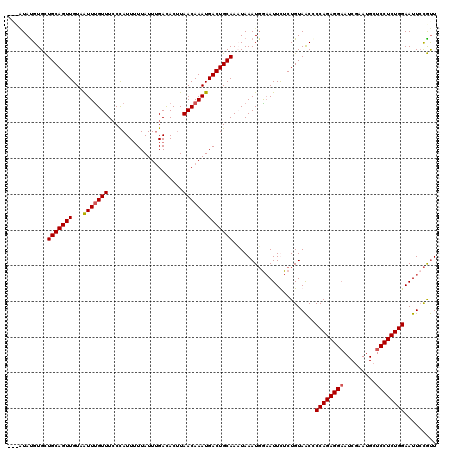
| Location | 6,892,834 – 6,892,954 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.03 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -25.04 |
| Energy contribution | -26.60 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6892834 120 - 22224390 AACGGAAUUCCAGAGGAGCAUUAGAUUCCUCUGGUGUUACAGAGAAUUUCAUUUAGUUGCAGUCAUUUGUUAAGUAUCAAAUAAAAAUGGUAAACAAAUUACAACUGCAGCACAUAUGUU ...((((((((((((((((....).))))))))(.....)...))))))).....((((((((.(((((((...(((((........))))))))))))....))))))))......... ( -32.80) >DroSec_CAF1 17163 117 - 1 AACGCAAUUCCAGAGGAGCAUUCGAUUCCUCUGGGGUUACAGAGAAUUCCAUUUAGUUGCAGUCAUUUGUUAGGUGUCAAAUAAAAAUAGGAAACAAAUUACAACUGCAGCACAUAU--- ...(.((((((((((((((....).))))))))))))).)...............((((((((.(((((........))))).......(....)........))))))))......--- ( -33.40) >DroEre_CAF1 16617 115 - 1 AACGGAAUUCCAGAGGAGCAUUCGAUUCCUCUGGGGUUACAGAAAAUUCCAUUUAUUUGCAGUCAUUUGUUAAGUGUCCAAUAA-AAUGGGAAACAAACUACAACUGCAGCACGUA---- .(((.((((((((((((((....).)))))))))))))..................(((((((..((((((.....((((....-..)))).)))))).....)))))))..))).---- ( -32.20) >DroYak_CAF1 19678 120 - 1 AGUAGAAUUCCAGAGGAGCAUUCGAUGCCUCUGGCGCUACAGAAAAUGCAAUUUAUUUGCAGUCAUUUGUUAAGUGUCAAAUAAAAAUGGGAAACGAAUUACAACUGCAACACAUACACA .((((....(((((((..(....)...)))))))..))))................(((((((.(((((........))))).......(....)........))))))).......... ( -27.90) >consensus AACGGAAUUCCAGAGGAGCAUUCGAUUCCUCUGGGGUUACAGAAAAUUCCAUUUAGUUGCAGUCAUUUGUUAAGUGUCAAAUAAAAAUGGGAAACAAAUUACAACUGCAGCACAUAU___ .....((((((((((((((....).))))))))))))).................((((((((.(((((........))))).......(....)........))))))))......... (-25.04 = -26.60 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:00 2006