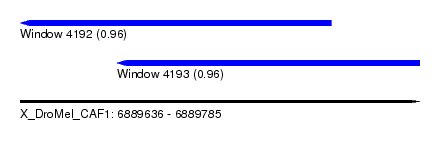
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,889,636 – 6,889,785 |
| Length | 149 |
| Max. P | 0.963959 |

| Location | 6,889,636 – 6,889,752 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6889636 116 - 22224390 CAAGUGCCGCGUUGGAGUCAGACAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCAUCCGCACCUCGUACAUAUCCAUAGCCGCAUAGCCUUGUC----CCAGGACACA ...((((((((.(((((((((......((((...((((((....)))))).))))...)))))))))..)))..............((..((......))..))..----...)).))). ( -32.80) >DroSec_CAF1 14066 114 - 1 CAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGACCACAUCCAUAGCCGCUU--CCUUGUC----CCAGGACACA ...(((.((.(((((((..((((((..((((...((((((....)))))).))))...))))))..))))).))..))..)))..............--...((((----....)))).. ( -36.00) >DroSim_CAF1 14769 114 - 1 CAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGACCACAUCCAUAGCCGAUU--CCUUGUC----CCAGGACACA ...(((.((.(((((((..((((((..((((...((((((....)))))).))))...))))))..))))).))..))..)))..............--...((((----....)))).. ( -36.00) >DroEre_CAF1 13385 102 - 1 CAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGUCCAUGUCC--------------UUGUC----CCAGGACACA ...((((......((((..((((((..((((...((((((....)))))).))))...))))))..))))))))........(((((--------------(....----..)))))).. ( -37.90) >DroYak_CAF1 16353 106 - 1 CAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCGGCACCUCCUUCACAUCC--------------UUGCCAGGCACAGGACACA ...(((((......(((..((((((..((((...((((((....)))))).))))...))))))..)))((((..............--------------.)))).)))))........ ( -39.16) >consensus CAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGUCCACAUCCAUAGCCG__U__CCUUGUC____CCAGGACACA ...(((((((...((((..((((((..((((...((((((....)))))).))))...))))))..))))))..............................((.......)))).))). (-29.20 = -29.80 + 0.60)

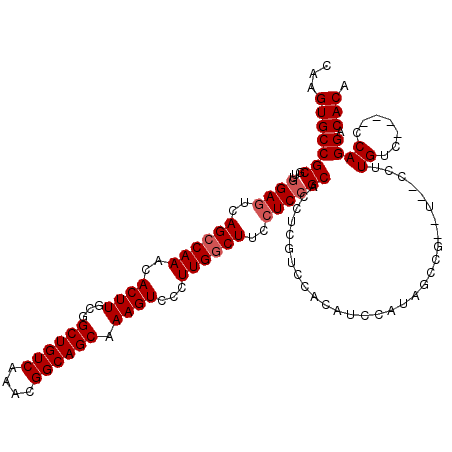
| Location | 6,889,672 – 6,889,785 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -34.04 |
| Energy contribution | -34.84 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6889672 113 - 22224390 ----CGGACCACUUUGCUUUGUCCGCCACUCG---AGGCUCAAGUGCCGCGUUGGAGUCAGACAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCAUCCGCACCUCGUA ----(((((...........))))).....((---((((......)))(((.(((((((((......((((...((((((....)))))).))))...)))))))))..)))...))).. ( -36.60) >DroSec_CAF1 14100 113 - 1 ----CGGACCACUUUGCUUUGUCCGCCACUCG---AGGCUCAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGAC ----(((((...........)))))....(((---((((......)))(((..(((...((((((..((((...((((((....)))))).))))...)))))))))..)))...)))). ( -42.00) >DroSim_CAF1 14803 113 - 1 ----CGGACCACUUUGCUUUGUCCGCCACUCG---AGGCUCAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGAC ----(((((...........)))))....(((---((((......)))(((..(((...((((((..((((...((((((....)))))).))))...)))))))))..)))...)))). ( -42.00) >DroEre_CAF1 13407 116 - 1 ----CGUACCACUUUGCUUUGUCCGCCACUCGACGAGGCUCAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGUC ----(((..(((((.((((((((........))))))))..)))))..)))..((((..((((((..((((...((((((....)))))).))))...))))))..)))).......... ( -45.30) >DroYak_CAF1 16379 117 - 1 GCCACGGACCACUUUGCUUUGUCCGCCACUCG---AGGCUCAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCGGCACCUCCUU (((.(((..(((((.((((((.........))---))))..)))))))).....(((..((((((..((((...((((((....)))))).))))...))))))..))))))........ ( -43.20) >consensus ____CGGACCACUUUGCUUUGUCCGCCACUCG___AGGCUCAAGUGCCGCGUUGGAGUCAGCCAAACACUUGCGGCUGUCAAACGGCAGCAAAGUCCCUUGGCUUCCUCCGCACCUCGUC .....((((...........))))((..........(((......))).....((((..((((((..((((...((((((....)))))).))))...))))))..))))))........ (-34.04 = -34.84 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:54 2006