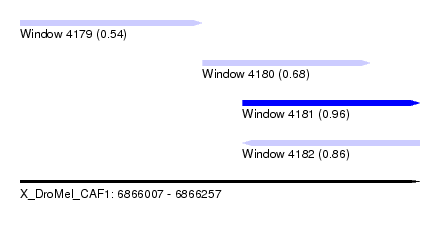
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,866,007 – 6,866,257 |
| Length | 250 |
| Max. P | 0.957620 |

| Location | 6,866,007 – 6,866,121 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6866007 114 + 22224390 GCCAAGUUUAUAAAACUAAAUUUAAACCAACAAAUCACUGUUCUACUUAAAUAACUUGAAACGAAGUGGUCGUUUCAGCUCCAUGCAGAUUUAAGUGAUUUCGUUCUGUUU-UUU ...(((((((......)))))))((((.(((((((((((..(((............((((((((.....))))))))((.....)))))....)))))))).)))..))))-... ( -23.30) >DroSec_CAF1 9726 115 + 1 GCCAAGUUAAUAAAACUAACUAUAAGCCAAACAAUCACUGUUUCACUCAAAUAACUUGAAACGAAGUGGUCAUUUCAGCUCCAUGCAGAUUUAAGUGAUUCAGUUCUGUUUUUUU ....(((((.......))))).......(((((...((((..(((((.((((..........(((((....))))).((.....))..)))).)))))..))))..))))).... ( -20.90) >DroSim_CAF1 10985 114 + 1 GCCAAGUCAAUAAAACUAACUAUAAGCCAAACAAUCACUGUUUUACUUAAAUAACUUGAAACGAAGUGGUCGUUUCAGCUCCAUGCAGAUUUAAGUGAUUCAGUUCUGUUU-UUU ............................(((((...((((..((((((((((....((((((((.....))))))))((.....))..))))))))))..))))..)))))-... ( -26.60) >consensus GCCAAGUUAAUAAAACUAACUAUAAGCCAAACAAUCACUGUUUUACUUAAAUAACUUGAAACGAAGUGGUCGUUUCAGCUCCAUGCAGAUUUAAGUGAUUCAGUUCUGUUU_UUU ....................................((((..((((((((((....((((((((.....))))))))((.....))..))))))))))..))))........... (-18.81 = -19.70 + 0.89)


| Location | 6,866,121 – 6,866,226 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6866121 105 + 22224390 CGUAAGUGCACU---------------UUCCACUUUUAAAUAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAGAUGGCUAACAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGA ..((((((((((---------------(((((..((.(((......))).))..)))...)))))((((((..(((((((...(((...)))...)))))))....))))))))))))). ( -31.10) >DroSec_CAF1 9841 103 + 1 UGUAAGUGCACU---------------UUCC--UUUUAAAUAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAAAUGGCUAACAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGA ..(((((((...---------------....--...........(....)....(((((.(((((.(((((..........)))))...))))).)))))............))))))). ( -27.00) >DroSim_CAF1 11099 103 + 1 UGUAAGUGCACU---------------UUCC--UUUUAAAUAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAAAUGGCUAACAGCACAGCUCUGGGCCUUCAUAAAUAGCCGUACUUGA ..(((((((...---------------....--...........(....).....((((.(((((.(((((..........)))))...))))).)))).............))))))). ( -25.80) >DroEre_CAF1 6768 100 + 1 ---AAGCGCAUU---------------UUUC--GCUUAAGUAGGGGCCUCACUUUGGCCAAGAGCGGCCGGCAAAUGGCCAAAAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGA ---(((((....---------------...)--))))(((((.((.........(((((.(((((.((.(((.....)))....))...))))).)))))..........)).))))).. ( -34.21) >DroYak_CAF1 6824 111 + 1 -------GCAUUUGCCUUGCUUUUUGAUUUC--CGUUAAAUAAUGCUUUCACUUUGGCCAAGAGUGGCCGUCAAAUGGCUAAAAGCACAGCUCUGAGCCAUCAUAAAUAGCUGUACUUGA -------((.((((((..((..((((((...--.))))))....)).........(((((....))))).......))).))).))((((((.(((....))).....))))))...... ( -25.20) >consensus _GUAAGUGCACU_______________UUCC__UUUUAAAUAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAAAUGGCUAACAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGA ..........................................................((((..(((((((...((((((...(((...)))...)))))).....)))))))..)))). (-20.58 = -20.50 + -0.08)

| Location | 6,866,146 – 6,866,257 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -26.95 |
| Energy contribution | -27.76 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
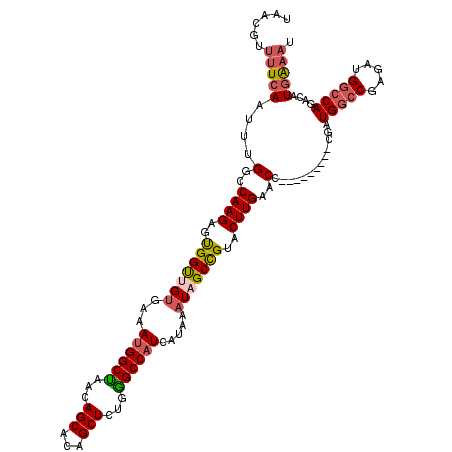
>X_DroMel_CAF1 6866146 111 + 22224390 UAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAGAUGGCUAACAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGAACC---------CGAUGGCCGAGAUGGCCAGACAUGAAAU ......(((((.((((((((......((((((...((.....)).))))))(((.(((((((......((.....))......---------.))))))).)))))))))))..))))). ( -39.12) >DroSec_CAF1 9864 102 + 1 UAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAAAUGGCUAACAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGAACC---------CGAUG---------GCCAGACAUGAAAU ......(((((.((((((((((..(((((((...((((((...(((...)))...)))))).....)))))))..))......---------...))---------))))))..))))). ( -30.80) >DroSim_CAF1 11122 111 + 1 UAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAAAUGGCUAACAGCACAGCUCUGGGCCUUCAUAAAUAGCCGUACUUGAACC---------CGAUGGCCGAGAUGGCCAGACAUGAAAU ......(((((.((((((((......((((((...((.....)).))))))(((.((((.((......((.....))......---------.)).)))).)))))))))))..))))). ( -34.82) >DroEre_CAF1 6788 111 + 1 UAGGGGCCUCACUUUGGCCAAGAGCGGCCGGCAAAUGGCCAAAAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGAACC---------CGCUGGCCGAGAUGGCCAGACAUGAAAU ..((((((.......)))((((.(((((......((((((...(((...)))...))))))........))))).))))..))---------)(((((((.....)))))).)....... ( -44.44) >DroYak_CAF1 6855 120 + 1 UAAUGCUUUCACUUUGGCCAAGAGUGGCCGUCAAAUGGCUAAAAGCACAGCUCUGAGCCAUCAUAAAUAGCUGUACUUGAACCGGAUGGCUGCGAUGGCCGAGAUGGCCAGACAUUGAAU .......((((.((((((((....((((((((..((((((...(((...)))...))))))......(((((((.((......))))))))).))))))))...))))))))...)))). ( -43.50) >consensus UAACGUUUUCAAUUUGGCCAAGAGUGGUUGUGAAAUGGCUAACAGCACAGCUCUGGGCCAUCAUAAAUAGCCGUACUUGAACC_________CGAUGGCCGAGAUGGCCAGACAUGAAAU ......(((((....(..((((..(((((((...((((((...(((...)))...)))))).....)))))))..))))..).............(((((.....)))))....))))). (-26.95 = -27.76 + 0.81)

| Location | 6,866,146 – 6,866,257 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -24.83 |
| Energy contribution | -25.04 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
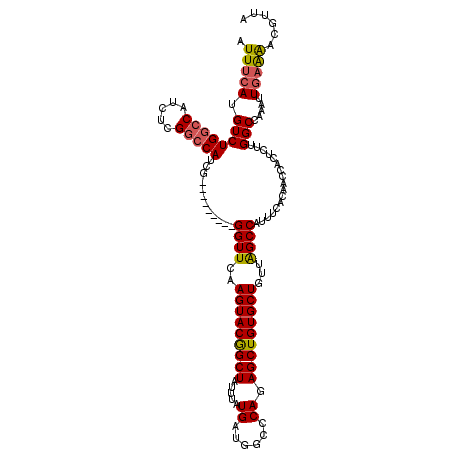
>X_DroMel_CAF1 6866146 111 - 22224390 AUUUCAUGUCUGGCCAUCUCGGCCAUCG---------GGUUCAAGUACGGCUAUUUAUGAUGGCCCAGAGCUGUGCUGUUAGCCAUCUCACAACCACUCUUGGCCAAAUUGAAAACGUUA .(((((....((((((....((((((((---------......((.....)).....))))))))..(((.((.((.....)))).)))...........))))))...)))))...... ( -33.20) >DroSec_CAF1 9864 102 - 1 AUUUCAUGUCUGGC---------CAUCG---------GGUUCAAGUACGGCUAUUUAUGAUGGCCCAGAGCUGUGCUGUUAGCCAUUUCACAACCACUCUUGGCCAAAUUGAAAACGUUA .(((((....((((---------((...---------((((.......(((((((...)))))))..(((.((.((.....)))).)))..)))).....))))))...)))))...... ( -28.60) >DroSim_CAF1 11122 111 - 1 AUUUCAUGUCUGGCCAUCUCGGCCAUCG---------GGUUCAAGUACGGCUAUUUAUGAAGGCCCAGAGCUGUGCUGUUAGCCAUUUCACAACCACUCUUGGCCAAAUUGAAAACGUUA .(((((....((((((....((......---------((((..(((((((((.....((......)).)))))))))...)))).........)).....))))))...)))))...... ( -30.36) >DroEre_CAF1 6788 111 - 1 AUUUCAUGUCUGGCCAUCUCGGCCAGCG---------GGUUCAAGUACGGCUAUUUAUGAUGGCCCAGAGCUGUGCUUUUGGCCAUUUGCCGGCCGCUCUUGGCCAAAGUGAGGCCCCUA .......(.((((((.....)))))))(---------((..((((..(((((......(((((((.(((((...))))).)))))))....)))))..))))(((.......)))))).. ( -47.30) >DroYak_CAF1 6855 120 - 1 AUUCAAUGUCUGGCCAUCUCGGCCAUCGCAGCCAUCCGGUUCAAGUACAGCUAUUUAUGAUGGCUCAGAGCUGUGCUUUUAGCCAUUUGACGGCCACUCUUGGCCAAAGUGAAAGCAUUA ....(((((.(((((.....)))))((((........((((.((((((((((.....(((....))).))))))))))..)))).......(((((....)))))...))))..))))). ( -42.60) >consensus AUUUCAUGUCUGGCCAUCUCGGCCAUCG_________GGUUCAAGUACGGCUAUUUAUGAUGGCCCAGAGCUGUGCUGUUAGCCAUUUCACAACCACUCUUGGCCAAAUUGAAAACGUUA .(((((.((((((((.....)))))............((((..(((((((((.....((......)).)))))))))...)))).................))).....)))))...... (-24.83 = -25.04 + 0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:44 2006