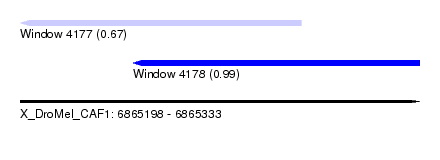
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,865,198 – 6,865,333 |
| Length | 135 |
| Max. P | 0.991274 |

| Location | 6,865,198 – 6,865,293 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6865198 95 - 22224390 AGAUCCA-------------AAAUUUGGGCGUCAG----------GACAUGCAAAUCUCGGUCAAUUGUGUGUCAAAGAUUGAUUGACAUGGCCGUGGAG--GCUGCGUUAAAUUUUUAG ......(-------------(((((((((((((((----------(((((((((...........))))))))).........)))))..((((.....)--))))).)))))))))... ( -22.60) >DroEre_CAF1 5948 92 - 1 AGACCCACU---------------UUGGGGGUCAG----------GACAUGCAAAUCUCGGUCGAUUGUGUGUCAAAGAUUGAUUGACAUGGCCGUGGAG--GCUGCGU-ACAAAUUUAG .(((((.(.---------------...))))))..----------(((((((((.((......))))))))))).........(((((..((((.....)--)))..))-.)))...... ( -28.50) >DroYak_CAF1 5862 119 - 1 AGAUCCACUUGGGAGAUCCCAUUUUUGGGGGUCAGCAAACAGGCUGACAUGCAAAUCUCGGCCAAUUGUGUGUCAAAGAUUGAUUGACAUGGCCGUGGAGUGGAGGCCU-ACAAAUUCAG ...(((((....(((((((((....)))))((((((......)))))).......))))(((((..(((..((((.....))))..)))))))))))))((((....))-))........ ( -44.10) >consensus AGAUCCACU___________A__UUUGGGGGUCAG__________GACAUGCAAAUCUCGGUCAAUUGUGUGUCAAAGAUUGAUUGACAUGGCCGUGGAG__GCUGCGU_ACAAAUUUAG .(((((......................))))).................(((..(((((((((..(((..((((.....))))..))))))))).))).....)))............. (-19.41 = -19.42 + 0.01)

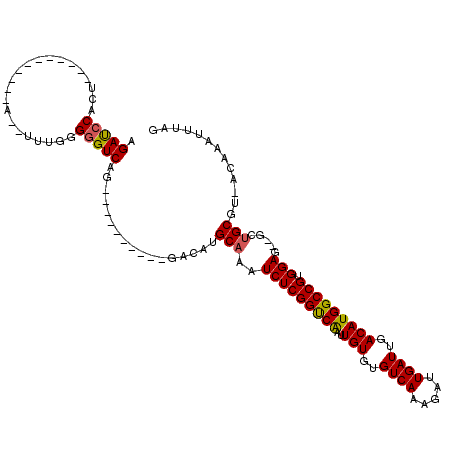
| Location | 6,865,236 – 6,865,333 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -22.18 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6865236 97 - 22224390 AUGUGAAUUAAUUUCAGGCACACGACCAUAUCCAUUUGGGAGAUCCA-------------AAAUUUGGGCGUCAG----------GACAUGCAAAUCUCGGUCAAUUGUGUGUCAAAGAU ...((((.....))))(((((((((((....((....))((((((((-------------.....)))((((...----------...))))..)))))))))....)))))))...... ( -27.60) >DroEre_CAF1 5985 95 - 1 AUGCGAAUUAAUUUCGGGCACACGACCACAUCCACUUGGCAGACCCACU---------------UUGGGGGUCAG----------GACAUGCAAAUCUCGGUCGAUUGUGUGUCAAAGAU ...((((.....))))(((((((((((.......((((((...(((...---------------..))).)))))----------)..((....))...))))....)))))))...... ( -28.20) >DroYak_CAF1 5901 120 - 1 AUGUGAAUUAAUUUCAGGCACACGACCAGCUCCACUUGGGAGAUCCACUUGGGAGAUCCCAUUUUUGGGGGUCAGCAAACAGGCUGACAUGCAAAUCUCGGCCAAUUGUGUGUCAAAGAU ...((((.....))))(((((((((((...((((..(((((..(((.....)))..)))))....))))))))........((((((.((....)).))))))....)))))))...... ( -44.40) >consensus AUGUGAAUUAAUUUCAGGCACACGACCACAUCCACUUGGGAGAUCCACU___________A__UUUGGGGGUCAG__________GACAUGCAAAUCUCGGUCAAUUGUGUGUCAAAGAU ...((((.....))))(((((((((((....((....))((((((((..((((....))))....)))).(((............))).......))))))))....)))))))...... (-22.18 = -23.10 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:40 2006