
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,861,750 – 6,861,902 |
| Length | 152 |
| Max. P | 0.996072 |

| Location | 6,861,750 – 6,861,868 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861750 118 - 22224390 GCCACUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUUGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCGUAGAUUCA--AGAAAACACCAGGCUGCGGCUUCUAAUUAU (((...(((((((((.((((((((....))))))))...((((......))))...)))))))))....((((((((((.........--.)))).....)))).))))).......... ( -35.40) >DroSec_CAF1 4638 118 - 1 GACAAUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUAGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCUUAGAUACA--AGAAAGCACCAGGCUGCGGCUUCUAAUUAU ........(((((((.((((((((....))))))))....(((......)))....)))))))(((((.((((..((((((......)--))))))))).)))))............... ( -34.20) >DroSim_CAF1 5025 118 - 1 GCCAAUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUUGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCUUAGAUCCA--AGAAGGCACCAGACUGCGGCUUCUAAUUAU (((.....(((((((.((((((((....))))))))...((((......))))...)))))))(((((.(((((((((..........--))))))))).)))))..))).......... ( -43.50) >DroEre_CAF1 4619 107 - 1 CCCAAUGUGAUUAAUCGUAGCUGCAUGUGCAGCUGCAGUCGCUCCGCAAGCCACUAGCUAAUCAGUUA-----------GUAGAUCCA--AGGAAGCCCCAAACUGCGCCCUCCAAUUAU ......((((((....((((((((....))))))))))))))..((((....(((((((....)))))-----------)).......--.((.....))....))))............ ( -29.70) >DroYak_CAF1 4604 112 - 1 CCCCAUUUGAUUAAUCGCUGCUGCAU----UGCUGCAUUUGGCUCGAAAGCCACUAGUUAAUCAUUUACGUUACUUUGCGUAGAUCCUUUACGUAGAUCCUAGUU----CUUCUAAUUAU .......((((((((.((.((.....----.)).))...(((((....)))))...))))))))..........((((((((((...))))))))))........----........... ( -23.60) >consensus GCCAAUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUUGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCGUAGAUCCA__AGAAAGCACCAGGCUGCGGCUUCUAAUUAU .......((((((((.((((((((....))))))))....(((......)))....))))))))........................................................ (-18.90 = -19.78 + 0.88)

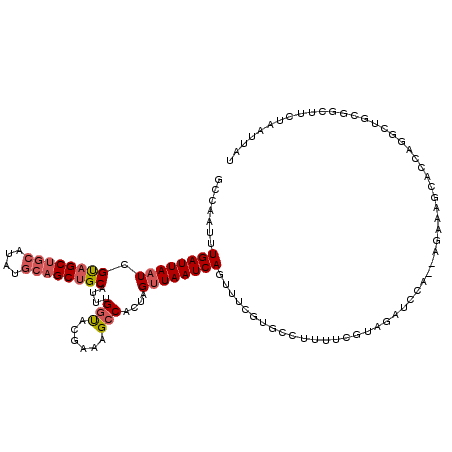
| Location | 6,861,788 – 6,861,902 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -20.96 |
| Energy contribution | -22.65 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861788 114 - 22224390 GCCAGCCAAUUUCCAGC-UGCCAUCCA---UUCUGU--AGGCCACUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUUGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCG (.((((.........))-)).).....---......--((((.((.(((((((((.((((((((....))))))))...((((......))))...)))))))))....))))))..... ( -33.30) >DroSec_CAF1 4676 114 - 1 GCCAGCCAAUUUCCUGC-UGCCAUUCA---UUCUGU--AGGACAAUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUAGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCU ......(((..((((((-.........---....))--))))....))).......((((((((....))))))))...(((((((((....(((........))))))))))))..... ( -33.02) >DroSim_CAF1 5063 114 - 1 GCCGGCCAAUUUCCUGC-UGCCACUCA---UUCUGU--AGGCCAAUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUUGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCU ...((((.....(((((-.........---....))--))).....(((((((((.((((((((....))))))))...((((......))))...)))))))))....).)))...... ( -34.02) >DroYak_CAF1 4640 116 - 1 GCCAGCCAAUUUCCUGCGUGGCAUGCAACAUAAUGGAUGCCCCCAUUUGAUUAAUCGCUGCUGCAU----UGCUGCAUUUGGCUCGAAAGCCACUAGUUAAUCAUUUACGUUACUUUGCG ((((((.........)).))))..((((..(((((((((....))))((((((((.((.((.....----.)).))...(((((....)))))...))))))))....)))))..)))). ( -30.70) >consensus GCCAGCCAAUUUCCUGC_UGCCAUUCA___UUCUGU__AGGCCAAUUUGAUUAAUCGUAGCUGCAUAUGCAGCUGCAUUUGGUACGAAAGCCACUAGUUAAUCAGUUUCGUGCCUUUUCG ..................................(...((((....(((((((((.((((((((....))))))))....(((......)))....)))))))))......))))...). (-20.96 = -22.65 + 1.69)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:39 2006