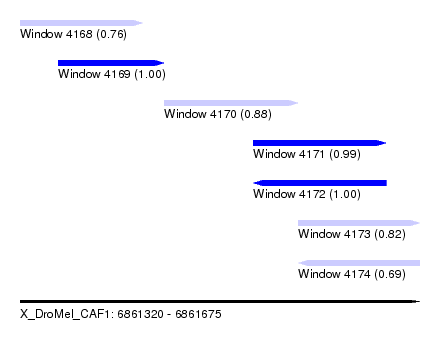
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,861,320 – 6,861,675 |
| Length | 355 |
| Max. P | 0.998776 |

| Location | 6,861,320 – 6,861,429 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.762061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861320 109 + 22224390 GACCGCCUGGGCAGACAGA--UGUAUG----UAUGUAUGUAAGUAAGUCUUGCUUGGCUAUAAUAAAGUGGCAUGACCAUGAAAGUG-----GCCAAACGGGGGGUCACUGUCAAAGGAA ((((.(((((((((((..(--(.((((----(....))))).))..))).))))(((((((......((((.....))))....)))-----))))..)))).))))............. ( -35.50) >DroSec_CAF1 4218 98 + 1 GGCCGCCUGGGCAGACAGC------------UAUGUAUGUAAGU-----UUGCCUGGCAAUAAUAAAGUGGCAUGGCCAUGAAAGUG-----GCCAAACGGGGGGUCACUGUCAAAGGAA (((.(((.((((((((.((------------.......))..))-----)))))))))........((((((.(((((((....)))-----))))..(....))))))))))....... ( -37.80) >DroEre_CAF1 4175 114 + 1 GGCCGCCUGGACAGACAGCAAAGUAAAAGUGUAUGUAUGUGAGU-----UUCUCGGGCAAUAAUAAAGUGGCAUGGCCAUGAAAGUGGCAUGGCCGAACCG-GGGUCACUGUCAAAGGAA ((((((((((..((((.(((..(((......)))...)))..))-----)).))))))..........((((...(((((....)))))...)))).....-.))))............. ( -34.20) >DroYak_CAF1 4189 93 + 1 GGCCGCCUGGGCAGCCAGCCAGCCA----------------AGU-----UUCCCUGGCCAUAAUCAAGUGGCAUGACCAUGAAAGUG-----UCCGCACGG-GGUUCACUGUCAAAGGAA (((.(.((((....)))).).))).----------------...-----...(((.(((((......))))).((((..((((.(((-----....)))..-..))))..)))).))).. ( -30.60) >consensus GGCCGCCUGGGCAGACAGC___GUA______UAUGUAUGUAAGU_____UUCCCUGGCAAUAAUAAAGUGGCAUGACCAUGAAAGUG_____GCCAAACGG_GGGUCACUGUCAAAGGAA .....(((.(((((..........................................(((((......))))).(((((......((...........))....))))))))))..))).. (-16.85 = -17.10 + 0.25)


| Location | 6,861,354 – 6,861,448 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -19.22 |
| Energy contribution | -20.33 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861354 94 + 22224390 AAGUAAGUCUUGCUUGGCUAUAAUAAAGUGGCAUGACCAUGAAAGUGGCCAAACGGGGGGUCACUGUCAAAGGAAUCCCUCGG------------AUAUAUCCCCC ...........(.((((((((......((((.....))))....)))))))).)(((((((...((((..(((....)))..)------------))).))))))) ( -31.10) >DroSec_CAF1 4246 89 + 1 AAGU-----UUGCCUGGCAAUAAUAAAGUGGCAUGGCCAUGAAAGUGGCCAAACGGGGGGUCACUGUCAAAGGAAUCCCUUGG------------AUAUAUCCCCC ..((-----(((((...............))))(((((((....))))))))))(((((((...((((.((((....)))).)------------))).))))))) ( -36.06) >DroYak_CAF1 4214 98 + 1 -AGU-----UUCCCUGGCCAUAAUCAAGUGGCAUGACCAUGAAAGUGUCCGCACGG-GGUUCACUGUCAAAGGAAUCCCUCUCCACUCCACUCGGAAAU-CCCCCC -.((-----((((...(((((......)))))........((..(((....)))((-(((((.((.....)))))))))............))))))))-...... ( -26.70) >consensus AAGU_____UUGCCUGGCAAUAAUAAAGUGGCAUGACCAUGAAAGUGGCCAAACGGGGGGUCACUGUCAAAGGAAUCCCUCGG____________AUAUAUCCCCC ..............(((((((......((((.....))))....)))))))..((((((((..((.....))..))))))))........................ (-19.22 = -20.33 + 1.12)



| Location | 6,861,448 – 6,861,567 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.16 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861448 119 + 22224390 UCUUGGCCAUCAGUAAGCGGCAAUCGUCGUCAUUAAUCAAACUGGCAACAGUAAGCCCCCGCAUUC-CCCAACCCUCGUCAACGAAAGCUUCUCGCUUUCGCUCUUUAUUGCAUUUGUCA ...((((.(((((((((.(((....(.((...........((((....))))..((....))....-.........)).)...((((((.....))))))))).))))))).))..)))) ( -26.10) >DroSec_CAF1 4335 119 + 1 UCUCGGCCAUCAGUAAGCGGCAAUCGUCGUCAUUAAUCAAACUGGCAACAGUAAGCCCCCGCAUUC-CCCAAUCCUCGUCAGCGAAAGCUUCUUGCUUUCGCUCUUUAUUGCAUUUGUCA ....(((.((((((((((((.....(....).........((((....))))......))))....-.............(((((((((.....)))))))))..)))))).))..))). ( -30.00) >DroSim_CAF1 4747 119 + 1 UCUCGGCCAUCAGUAAGCGGCAAUCGUCGUCAUUAAUCAAACUGGCAACAGUAAGCCCCCGCAUUC-CCCAACCCUCGUCAGCGAAAGCUUCUCGCUUUCGCUCUUUAUUGCAUUUGUCA ....(((.((((((((((((.....(....).........((((....))))......))))....-.............(((((((((.....)))))))))..)))))).))..))). ( -29.40) >DroEre_CAF1 4326 119 + 1 UUUUGGCCAUCAGUC-GCGGCAAUCGUAGUCAUUAAUCAAACUGGCAACAGUAUGCCCCCACAGAUCCCCAACCCUCGUCAACGAAAGCGUCUCGCUUUUGCUCUUUAUUGCAUUUGUCA ..((((..(((.((.-(.((((...(....).........((((....)))).)))).).)).)))..)))).....(.(((.(((((((...)))))))((........))..))).). ( -25.10) >DroYak_CAF1 4312 118 + 1 UCUCGGCCAUCAGUG-GCGGCAAUCGUCGUCAUUAAUCAAACUGGCUACAGUAGGCCCCCACACUU-CGCAACCCUCGUCAAGGAGAGCUUCUCGCUUUUGCUCUUUAUUGCAUUUGUCA ....((((...((((-(((((....)))))))))......((((....)))).)))).........-.((((.(((.....)))(((((...........)))))...))))........ ( -33.90) >consensus UCUCGGCCAUCAGUAAGCGGCAAUCGUCGUCAUUAAUCAAACUGGCAACAGUAAGCCCCCGCAUUC_CCCAACCCUCGUCAACGAAAGCUUCUCGCUUUCGCUCUUUAUUGCAUUUGUCA ....(((.........(((((....)))))..........((((....))))..)))....................(.(((.((((((.....))))))((........))..))).). (-23.60 = -23.16 + -0.44)


| Location | 6,861,527 – 6,861,645 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.07 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -11.76 |
| Energy contribution | -12.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861527 118 + 22224390 AACGAAAGCUUCUCGCUUUCGCUCUUUAUUGCAUUUGUCACCACAGCAUGAACAAUGCACUGUUCGAAAAAU-UGUAUGUUUUCACUUCUUUCACACAGUUUAAU-CGUGAAAACACAUA ..(((((((.....)))))))........((((((..(((........)))..)))))).............-(((.(((((((((...................-.))))))))).))) ( -23.35) >DroSec_CAF1 4414 119 + 1 AGCGAAAGCUUCUUGCUUUCGCUCUUUAUUGCAUUUGUCACCACAGCAUGAACAGUGCACUGUUCGAAAAAU-UGUACGUUUUCACUUCUUUCACGCAGUUUAAUGUGUGAAUACCCAUA (((((((((.....)))))))))......(((...(((....)))))).((((((....))))))(((((..-......)))))......((((((((......))))))))........ ( -33.60) >DroSim_CAF1 4826 118 + 1 AGCGAAAGCUUCUCGCUUUCGCUCUUUAUUGCAUUUGUCACCACAGCAUGAACAGUGCACUGUUCGAAAAAU-UGGACGUUUUCACUUCUUUCACGCAGUUUAAU-CAUGAAUACGCCUA (((((((((.....)))))))))....(((((.................((((((....))))))((((((.-((((....)))).)).))))..))))).....-.............. ( -27.40) >DroEre_CAF1 4405 96 + 1 AACGAAAGCGUCUCGCUUUUGCUCUUUAUUGCAUUUGUCACCACAGCAUGGA---UGCACUGGGCGAAACAA-GUGAUGGCUUCAUCACCC--------------------ACACACACU ...(((((((...)))))))((((.....((((((..((......).)..))---))))..)))).......-((((((....))))))..--------------------......... ( -24.50) >DroYak_CAF1 4390 94 + 1 AAGGAGAGCUUCUCGCUUUUGCUCUUUAUUGCAUUUGUCACCACAGCACAGA---UGCACAGGGCGAAAAAAUGCUAUGGCUUCA---ACU--------------------AUACACAUA .((..(((((....(((((((((((....((((((((((......).)))))---)))).)))))))))....))...)))))..---.))--------------------......... ( -24.70) >consensus AACGAAAGCUUCUCGCUUUCGCUCUUUAUUGCAUUUGUCACCACAGCAUGAACA_UGCACUGUUCGAAAAAU_UGUAUGUUUUCACUUCUUUCAC_CAGUUUAAU___UGAAUACACAUA ...((((((.....))))))((........)).............((((.....)))).......(((((.........))))).................................... (-11.76 = -12.04 + 0.28)


| Location | 6,861,527 – 6,861,645 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Mean single sequence MFE | -32.61 |
| Consensus MFE | -17.58 |
| Energy contribution | -19.62 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861527 118 - 22224390 UAUGUGUUUUCACG-AUUAAACUGUGUGAAAGAAGUGAAAACAUACA-AUUUUUCGAACAGUGCAUUGUUCAUGCUGUGGUGACAAAUGCAAUAAAGAGCGAAAGCGAGAAGCUUUCGUU ..(((((((((((.-...................)))))))))))..-.......((((((....)))))).(((..((....))...)))......(((((((((.....))))))))) ( -34.75) >DroSec_CAF1 4414 119 - 1 UAUGGGUAUUCACACAUUAAACUGCGUGAAAGAAGUGAAAACGUACA-AUUUUUCGAACAGUGCACUGUUCAUGCUGUGGUGACAAAUGCAAUAAAGAGCGAAAGCAAGAAGCUUUCGCU .....((((((((.((......)).)))).....((....)))))).-.......((((((....)))))).(((..((....))...)))......(((((((((.....))))))))) ( -36.10) >DroSim_CAF1 4826 118 - 1 UAGGCGUAUUCAUG-AUUAAACUGCGUGAAAGAAGUGAAAACGUCCA-AUUUUUCGAACAGUGCACUGUUCAUGCUGUGGUGACAAAUGCAAUAAAGAGCGAAAGCGAGAAGCUUUCGCU ..(((((.(((((.-.((..((...))..))...))))).)))))..-.......((((((....)))))).(((..((....))...)))......(((((((((.....))))))))) ( -37.80) >DroEre_CAF1 4405 96 - 1 AGUGUGUGU--------------------GGGUGAUGAAGCCAUCAC-UUGUUUCGCCCAGUGCA---UCCAUGCUGUGGUGACAAAUGCAAUAAAGAGCAAAAGCGAGACGCUUUCGUU ..((((((.--------------------.(((((((....))))))-)..).(((((((((...---.....)))).)))))...)))))......(((.((((((...)))))).))) ( -29.30) >DroYak_CAF1 4390 94 - 1 UAUGUGUAU--------------------AGU---UGAAGCCAUAGCAUUUUUUCGCCCUGUGCA---UCUGUGCUGUGGUGACAAAUGCAAUAAAGAGCAAAAGCGAGAAGCUCUCCUU ....(((((--------------------..(---((..((((((((((.....(((...)))..---...))))))))))..))))))))....(((((...........))))).... ( -25.10) >consensus UAUGUGUAUUCA___AUUAAACUG_GUGAAAGAAGUGAAAACAUACA_AUUUUUCGAACAGUGCA_UGUUCAUGCUGUGGUGACAAAUGCAAUAAAGAGCGAAAGCGAGAAGCUUUCGUU ....................................(((((.........)))))((((((....)))))).(((..((....))...)))......(((((((((.....))))))))) (-17.58 = -19.62 + 2.04)


| Location | 6,861,567 – 6,861,675 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -15.50 |
| Energy contribution | -17.50 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861567 108 + 22224390 CCACAGCAUGAACAAUGCACUGUUCGAAAAAUUGUAUGUUUUCACUUCUUUCACACAGUUUAAU-CGUGAAAACACAUAACUUAGUUCUAUCAUUUUCUCGCUGUGCAC .((((((..(((((......)))))(((...((((.(((((((((...................-.))))))))).)))).....)))............))))))... ( -25.05) >DroSec_CAF1 4454 109 + 1 CCACAGCAUGAACAGUGCACUGUUCGAAAAAUUGUACGUUUUCACUUCUUUCACGCAGUUUAAUGUGUGAAUACCCAUAACAUAGGUCUACCAUUUUCUCGCUGUGCAA .((((((..((((((....))))))(((((.......(((.........((((((((......)))))))).......)))...((....)).)))))..))))))... ( -32.19) >DroSim_CAF1 4866 99 + 1 CCACAGCAUGAACAGUGCACUGUUCGAAAAAUUGGACGUUUUCACUUCUUUCACGCAGUUUAAU-CAUGAAUACGCCUAACAUAGGUCUACCAUUA---------GAAA ((......(((((((....))))))).......((.(((.((((....................-..)))).))))).......))((((....))---------)).. ( -15.45) >consensus CCACAGCAUGAACAGUGCACUGUUCGAAAAAUUGUACGUUUUCACUUCUUUCACGCAGUUUAAU_CGUGAAUACACAUAACAUAGGUCUACCAUUUUCUCGCUGUGCAA .((((((.(((((((....))))))).....((((.(((.(((((.....................))))).))).))))....................))))))... (-15.50 = -17.50 + 2.00)


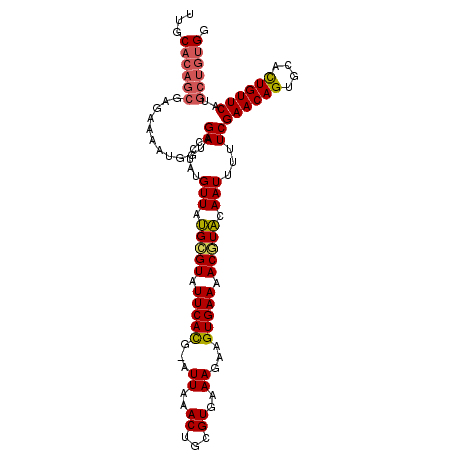
| Location | 6,861,567 – 6,861,675 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -19.10 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6861567 108 - 22224390 GUGCACAGCGAGAAAAUGAUAGAACUAAGUUAUGUGUUUUCACG-AUUAAACUGUGUGAAAGAAGUGAAAACAUACAAUUUUUCGAACAGUGCAUUGUUCAUGCUGUGG ...(((((((...........(((..(((((.(((((((((((.-...................))))))))))).))))))))((((((....)))))).))))))). ( -30.25) >DroSec_CAF1 4454 109 - 1 UUGCACAGCGAGAAAAUGGUAGACCUAUGUUAUGGGUAUUCACACAUUAAACUGCGUGAAAGAAGUGAAAACGUACAAUUUUUCGAACAGUGCACUGUUCAUGCUGUGG ...((((((((((((.((....((((((...)))))).(((((.((......)).)))))....((....))...)).))))))((((((....))))))..)))))). ( -34.90) >DroSim_CAF1 4866 99 - 1 UUUC---------UAAUGGUAGACCUAUGUUAGGCGUAUUCAUG-AUUAAACUGCGUGAAAGAAGUGAAAACGUCCAAUUUUUCGAACAGUGCACUGUUCAUGCUGUGG ....---------..((((((((.....(((.(((((.(((((.-.((..((...))..))...))))).))))).)))...))((((((....)))))).)))))).. ( -20.60) >consensus UUGCACAGCGAGAAAAUGGUAGACCUAUGUUAUGCGUAUUCACG_AUUAAACUGCGUGAAAGAAGUGAAAACGUACAAUUUUUCGAACAGUGCACUGUUCAUGCUGUGG ...((((((............((.....(((.(((((.(((((...((..((...))..))...))))).))))).)))...))((((((....))))))..)))))). (-19.10 = -20.00 + 0.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:37 2006