
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,830,258 – 6,830,387 |
| Length | 129 |
| Max. P | 0.725557 |

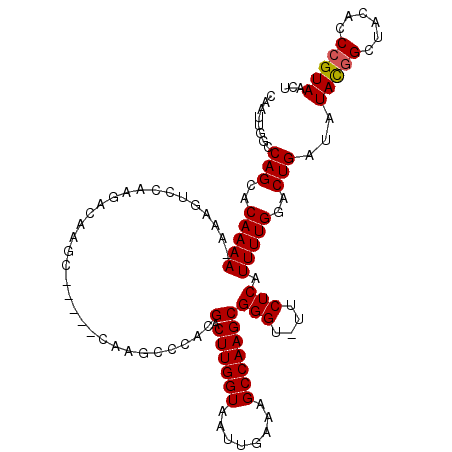
| Location | 6,830,258 – 6,830,366 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.01 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6830258 108 - 22224390 CAAUUGGCCAGCACAAAA-AAAGGCCAAGACAAGC-----CAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU-UUCUCGUUUUGGACUGAUAUAUGGCUACACCCGUAACU ....((((((........-..((.(((((((.((.-----.((((((..(.((((((........)))))))))))-)))).))))))).))......))))))........... ( -30.89) >DroSec_CAF1 10484 113 - 1 CAAUUGGCCAGCACAAAA-AAAGUCCAAGACAAGCCAAGCCAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU-UUCUCAUUUUGGACUGAUAUACGGCUACACCCGUAACU ...(((.......)))..-..(((((((((...((((((.(........).))))))....((((.((....)).)-)))...)))))))))....(((((......)))))... ( -28.80) >DroSim_CAF1 10505 114 - 1 CAAUUGGCCAGCACAAAAAAAAGUCCAAGACAAGCCAAGCCAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU-UUCUCAUUUUGGACUGAUAUACGGCUACACCCGUAACU ...(((.......))).....(((((((((...((((((.(........).))))))....((((.((....)).)-)))...)))))))))....(((((......)))))... ( -28.80) >DroYak_CAF1 10753 91 - 1 CAAUUGGCCAGCACAAAA------------------------AGCCCAAGACUUGGUAAUUGAAAGCCAAGCGGGUUUUCUCAUUUUGGACUGAUAUAUUGCUACACCCGUAACU ..((..(((((.....((------------------------(((((..(.((((((........))))))))))))))......)))).)..))...((((.......)))).. ( -20.50) >consensus CAAUUGGCCAGCACAAAA_AAAGUCCAAGACAAGC_____CAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU_UUCUCAUUUUGGACUGAUAUACGGCUACACCCGUAACU ........(((..(((((...............................(.((((((........)))))))(((....))).)))))..)))...(((((......)))))... (-18.48 = -18.48 + -0.00)

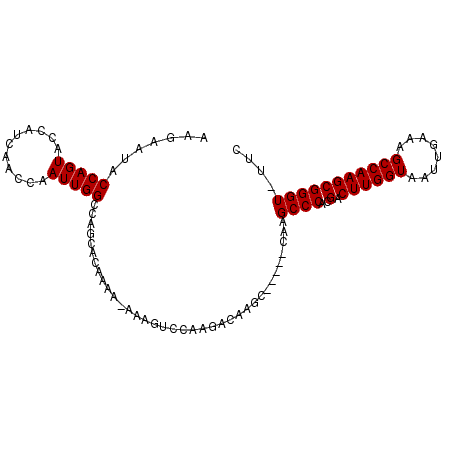
| Location | 6,830,293 – 6,830,387 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6830293 94 - 22224390 AAGUAUACCAGUACCAUCAACCAAUUGGCCAGCACAAAA-AAAGGCCAAGACAAGC-----CAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU-UUC ..((((....))))..........((((((.........-...)))))).......-----.((((((..(.((((((........)))))))))))-)). ( -22.30) >DroSec_CAF1 10519 99 - 1 AAGAAUACCAGUACCAUCAACCAAUUGGCCAGCACAAAA-AAAGUCCAAGACAAGCCAAGCCAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU-UUC ..................((((..(((((...((.....-...(((...)))..((((((.(........).))))))...))...)))))...)))-).. ( -19.70) >DroSim_CAF1 10540 100 - 1 AAGAAUACCAGUACCAUCAACCAAUUGGCCAGCACAAAAAAAAGUCCAAGACAAGCCAAGCCAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU-UUC ..................((((..(((((...((.........(((...)))..((((((.(........).))))))...))...)))))...)))-).. ( -19.70) >DroYak_CAF1 10788 77 - 1 AAGUAUACCAGUACCGCCAACCAAUUGGCCAGCACAAAA------------------------AGCCCAAGACUUGGUAAUUGAAAGCCAAGCGGGUUUUC ..((((....)))).(((((....)))))........((------------------------(((((..(.((((((........)))))))))))))). ( -20.10) >consensus AAGAAUACCAGUACCAUCAACCAAUUGGCCAGCACAAAA_AAAGUCCAAGACAAGC_____CAAGCCCACGACUUGGUAAUUGAAAGCCAAGCGGGU_UUC .......(((((...........)))))....................................((((..(.((((((........))))))))))).... (-13.73 = -13.72 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:25 2006