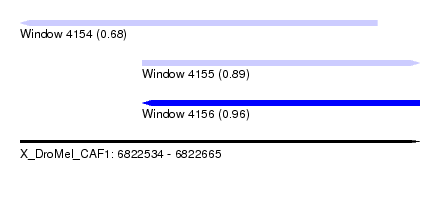
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,822,534 – 6,822,665 |
| Length | 131 |
| Max. P | 0.957501 |

| Location | 6,822,534 – 6,822,651 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -31.65 |
| Energy contribution | -31.27 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6822534 117 - 22224390 GUGCC---AAGAUGUAUAAGUUGCUGGGUAGCCUGAAGAGCUACCUCAAGUGGCAGGACAUCCAGACGGACAACGCCGUCUUCCGGCUGCACAACUCCUUUACCACGGUGCUCCUGCUAA ..(((---..(((((....((..((((((((((....).)))))))..))..))...))))).((((((......))))))...))).((((...............))))......... ( -38.06) >DroVir_CAF1 19226 117 - 1 GUGCC---AAGAUGUAUAAGCUGUUGGGUAGCCUGAAGAGCUACCUCAAGUGGCAGGACAUCCAGACGGAUAACGCGGUCUUCCGUCUGCACAACUCCUUUACAACCGUACUGCUGCUCA ((((.---..(((((....((..((((((((((....).)))))))..))..))...)))))((((((((..((...))..))))))))..................))))......... ( -36.10) >DroGri_CAF1 3736 117 - 1 GUGCC---AAGAUGUAUAAGCUAUUGGGUAGCCUAAAGAGCUACCUAAAGUGGCAGGACAUCCAGACGGACAAUGCCGUCUUCCGUCUGCACAACUCCUUCACAACGGUACUGCUGCUCA (((((---..(((((....((((((((((((((....).))))))))..)))))...)))))((((((((...........)))))))).................)))))......... ( -40.40) >DroWil_CAF1 3902 120 - 1 GUGCCAACAAGAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUUAAGUGGCAGGACAUUCAAACGGAUAAUGCCGUCUUUCGUUUGCAUAAUUCAUUCACCACGGUCCUUUUGCUGA ((((.(((..(((((....((((((((((((((....).)))))))..))))))...)))))...((((......)))).....))).)))).............((((......)))). ( -31.40) >DroMoj_CAF1 7565 117 - 1 GUGCC---AAAAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUCAAAUGGCAGGACAUCCAGACGGAUAAUGCAGUCUUCCGGCUCCACAACUCCUUCACAACUGUACUUUUGCUUA ..((.---((((.(((((.((((((((((((((....).)))))))..))))))((((.((((....))))..((.((((....)))).))....)))).......)))))))))))... ( -31.80) >DroAna_CAF1 3785 117 - 1 GUGCC---AAGAUGUACAAGUUACUGGGUAGCCUGAAGAGCUACCUAAAGUGGCAGGACAUCCAGACGGACAAUGCCGUCUUCCGUCUCCACAACUCCUUCACCACGGUGCUCCUGCUCA (..((---..(.((....((((..(((((((((....).))))))))..((((..((((....((((((......))))))...))))))))))))....)).)..))..)......... ( -36.80) >consensus GUGCC___AAGAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUCAAGUGGCAGGACAUCCAGACGGACAAUGCCGUCUUCCGUCUGCACAACUCCUUCACAACGGUACUCCUGCUCA (((((.....(((((....((((((((((((((....).)))))))..))))))...)))))((((((((..((...))..)))))))).................)))))......... (-31.65 = -31.27 + -0.38)

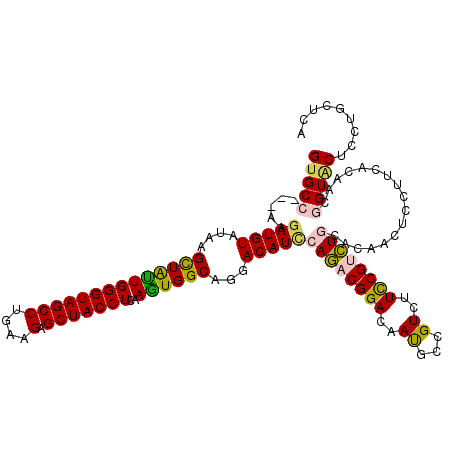
| Location | 6,822,574 – 6,822,665 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.62 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6822574 91 + 22224390 GACGGCGUUGUCCGUCUGGAUGUCCUGCCACUUGAGGUAGCUCUUCAGGCUACCCAGCAACUUAUACAUCUU---GGCACAGUU-------UG-----GAUCUC-------A--G----- (((((......)))))..((.(((((((((.(((.(((((((.....))))))))))..............)---)))).....-------.)-----))).))-------.--.----- ( -25.96) >DroVir_CAF1 19266 90 + 1 GACCGCGUUAUCCGUCUGGAUGUCCUGCCACUUGAGGUAGCUCUUCAGGCUACCCAACAGCUUAUACAUCUU---GGCACUAUU-------GU------------------AUUAGUU-- .............(((.((((((...((...(((.(((((((.....))))))))))..))....)))))).---)))((((..-------..------------------..)))).-- ( -25.20) >DroGri_CAF1 3776 108 + 1 GACGGCAUUGUCCGUCUGGAUGUCCUGCCACUUUAGGUAGCUCUUUAGGCUACCCAAUAGCUUAUACAUCUU---GGCACAGUU-------GAAGCGGGAUCUGGUACAGAACUAGUU-- (((((......))))).....(((((((.......(((((((.....)))))))((((.(((..........---)))...)))-------)..)))))))(((((.....)))))..-- ( -32.00) >DroWil_CAF1 3942 97 + 1 GACGGCAUUAUCCGUUUGAAUGUCCUGCCACUUAAGGUAGCUCUUCAGGCUACCCAAUAGCUUAUACAUCUUGUUGGCACACUACUUAAUUAC-----GAUC------------------ ..((..((((...(((((((.(..(((((......)))))..))))))))...(((((((..........))))))).........))))..)-----)...------------------ ( -19.20) >DroMoj_CAF1 7605 88 + 1 GACUGCAUUAUCCGUCUGGAUGUCCUGCCAUUUGAGGUAGCUCUUCAGGCUACCCAAUAGCUUAUACAUUUU---GGCACAAUU-------UC------------------A--AAUU-- .............(((..(((((...((...(((.(((((((.....))))))))))..))....)))))..---)))......-------..------------------.--....-- ( -22.40) >DroPer_CAF1 3331 96 + 1 GACGGCGUUGUCCGUCUGGAUGUCCUGCCAUUUGAGGUAGCUCUUCAGGCUACCCAAUAGCUUAUACAUCUU---GCCACAGUU-------UG-----GAUCUC-------A--AAUUUG (((((......))))).((((((...((...(((.(((((((.....))))))))))..))....)))))).---.....((((-------((-----.....)-------)--)))).. ( -27.60) >consensus GACGGCAUUAUCCGUCUGGAUGUCCUGCCACUUGAGGUAGCUCUUCAGGCUACCCAAUAGCUUAUACAUCUU___GGCACAGUU_______UC_____GAUC_________A__A_UU__ (((((......))))).((((((...((...(((.(((((((.....))))))))))..))....))))))................................................. (-19.00 = -19.62 + 0.61)

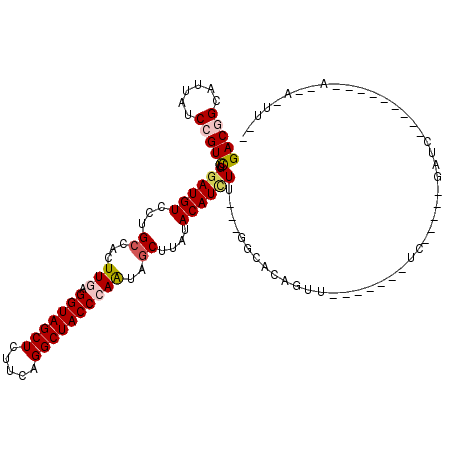

| Location | 6,822,574 – 6,822,665 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
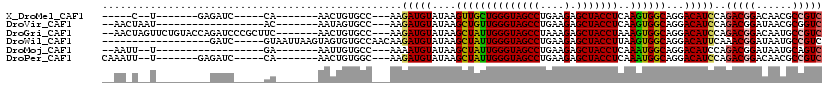
>X_DroMel_CAF1 6822574 91 - 22224390 -----C--U-------GAGAUC-----CA-------AACUGUGCC---AAGAUGUAUAAGUUGCUGGGUAGCCUGAAGAGCUACCUCAAGUGGCAGGACAUCCAGACGGACAACGCCGUC -----.--.-------((..((-----(.-------...(((((.---.....))))).((..((((((((((....).)))))))..))..)).)))..))..(((((......))))) ( -29.60) >DroVir_CAF1 19266 90 - 1 --AACUAAU------------------AC-------AAUAGUGCC---AAGAUGUAUAAGCUGUUGGGUAGCCUGAAGAGCUACCUCAAGUGGCAGGACAUCCAGACGGAUAACGCGGUC --.((((..------------------..-------..))))(((---..(((((....((..((((((((((....).)))))))..))..))...)))))..(.((.....)))))). ( -24.50) >DroGri_CAF1 3776 108 - 1 --AACUAGUUCUGUACCAGAUCCCGCUUC-------AACUGUGCC---AAGAUGUAUAAGCUAUUGGGUAGCCUAAAGAGCUACCUAAAGUGGCAGGACAUCCAGACGGACAAUGCCGUC --.....(((((((..(.......(((((-------(.((.....---.)).))...))))..((((((((((....).))))))))).)..))))))).....(((((......))))) ( -32.20) >DroWil_CAF1 3942 97 - 1 ------------------GAUC-----GUAAUUAAGUAGUGUGCCAACAAGAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUUAAGUGGCAGGACAUUCAAACGGAUAAUGCCGUC ------------------....-----........(.(((((.((.((.....))....((((((((((((((....).)))))))..)))))).))))))))..((((......)))). ( -25.20) >DroMoj_CAF1 7605 88 - 1 --AAUU--U------------------GA-------AAUUGUGCC---AAAAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUCAAAUGGCAGGACAUCCAGACGGAUAAUGCAGUC --....--.------------------..-------.........---....(((((..((((((((((((((....).)))))))..)))))).....((((....)))).)))))... ( -23.00) >DroPer_CAF1 3331 96 - 1 CAAAUU--U-------GAGAUC-----CA-------AACUGUGGC---AAGAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUCAAAUGGCAGGACAUCCAGACGGACAACGCCGUC ((..((--(-------(.....-----))-------))...))..---..(((((....((((((((((((((....).)))))))..))))))...)))))..(((((......))))) ( -28.20) >consensus __AA_U__U_________GAUC_____CA_______AACUGUGCC___AAGAUGUAUAAGCUAUUGGGUAGCCUGAAGAGCUACCUCAAGUGGCAGGACAUCCAGACGGACAACGCCGUC ..................................................(((((....((((((((((((((....).)))))))..))))))...)))))..(((((......))))) (-23.46 = -23.30 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:21 2006