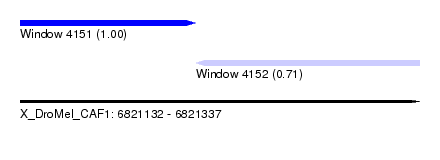
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,821,132 – 6,821,337 |
| Length | 205 |
| Max. P | 0.995269 |

| Location | 6,821,132 – 6,821,222 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6821132 90 + 22224390 AACCGAGAUAGGUGUAAAUUUGAAAUUUACUCACACCAUAGAGAUAUGAUAAUUGUAUGUGCAAUUGAAACAUGUGAUUAUCAAGUUUCA----------------------------- .(((......)))((((((.....))))))..........(((((.(((((((..(((((.........)))))..))))))).))))).----------------------------- ( -23.80) >DroSec_CAF1 1503 119 + 1 AAACGAGAUAGGUGUAAAUUUGAGAUUUACUCACACCACAGAGAUAUGAUAAUUGUAUGUGCAAUUGAUAUGUGUGAUUACCAAGUUUCAGAUCAACAUCUUUCUGUCUUUAUAACACA .....(((.((((((..(((((((((((..((((((..((......)).(((((((....)))))))....)))))).....)))))))))))..)))))).))).............. ( -28.40) >DroSim_CAF1 1517 119 + 1 AAACGAGAUAGGUGUAAAUUUGAGAUUUGCUCACACCAUAGAGAUAUGAUAAUUGUAUGUGCAAUUGAUAUGCGUGAUUACCAAGUUUCAGAUCAACAUCUUUCUGUCUUUAUAAUACA .....(((.((((((..((((((((((((.((((..((((....)))).(((((((....)))))))......))))....))))))))))))..)))))).))).............. ( -30.50) >consensus AAACGAGAUAGGUGUAAAUUUGAGAUUUACUCACACCAUAGAGAUAUGAUAAUUGUAUGUGCAAUUGAUAUGUGUGAUUACCAAGUUUCAGAUCAACAUCUUUCUGUCUUUAUAA_ACA ....(((((.((((((((((...)))))))((((((((((....)))).(((((((....)))))))....))))))..)))..))))).............................. (-16.56 = -16.90 + 0.34)

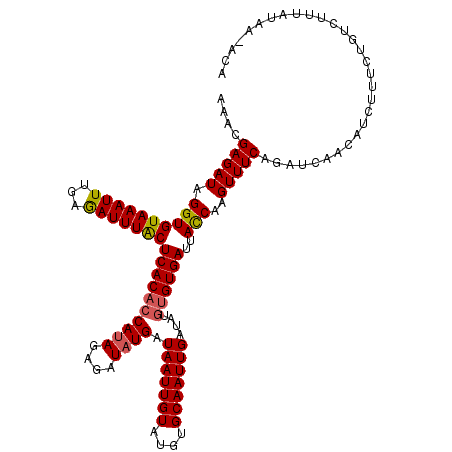
| Location | 6,821,222 – 6,821,337 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -20.12 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6821222 115 - 22224390 GGCCAAUGAUUUUGGCGACGAGGAUGCCAAGAAGUACUACACCUACUACCAGUGGGUGUGCUUCGUGCUUUUCUUCCAGGUGAGUUUCGUAGCA--UAAGAUUUUACUUCUAGAUUC .(((((.....))))).............(((((((..((((((((.....))))))))(((.((.((((.((.....)).))))..)).))).--........)))))))...... ( -33.90) >DroEre_CAF1 1777 107 - 1 GGCCAAUGACUUUGGCGAUGAGGAUGCCAAGAAGUACUACACCUACUAUCAGUGGGUGUGCUUCGUGCUUUUCUUCCAGGUGAGUUUCGCAUUUACUUCUA---C-------GAUUC .(((((.....)))))...(((((.((...(((((...((((((((.....)))))))))))))..))...))))).((((((((.....))))))))...---.-------..... ( -33.90) >DroWil_CAF1 1936 96 - 1 GGCCAAUGAUUUUGAUGACGAAGAUGCCAAGAAAUAUUAUACCUACUAUCAAUGGGUUUGCUUUGUGCUCUUCUUUCAGGUAAGUGUCGGCGGA--GA------------------- .(((.((.((((...(((.(((((.((((((.........(((((.......)))))....)))).)))))))..)))...)))))).)))...--..------------------- ( -18.12) >DroYak_CAF1 1643 105 - 1 GGCCAAUGAUUUUGGCGAUGAGGAUGCCAAGAAGUACUACACCUACUACCAGUGGGUCUGCUUCGUGCUUUUCUUCCAGGUGAGUUUCCUAUCU--UAAGG---C-------ACUUC .(((((.....)))))...((((.((((..((((((....((((((.....)))))).))))))..((((.((.....)).)))).........--...))---)-------))))) ( -31.10) >DroAna_CAF1 2282 96 - 1 GGCCAAUGACUUUGGUGACGAAGAUGCCAAGAAAUACUACACCUACUACCAAUGGGUGUGCUUUGUGCUCUUCUUCCAGGUGAGUUAUAUA---------------CUCCU------ .(((((.....)))))((.(((((.((((((......((((((((.......)))))))).)))).))))))).))..((.((((.....)---------------)))))------ ( -24.90) >DroPer_CAF1 1625 95 - 1 GUCCAAUGAUUUCGGAGACGAGGAUGCCAAGAAAUACUAUACCUACUACCAGUGGGUAUGCUUUGUGCUCUUCUUUCAGGUGAGUAA-AUA---------------GUCUG------ ((((.......(((....)))))))....(((.....(((((((((.....)))))))))((...(((((.((.....)).))))).-..)---------------)))).------ ( -23.91) >consensus GGCCAAUGAUUUUGGCGACGAGGAUGCCAAGAAAUACUACACCUACUACCAGUGGGUGUGCUUCGUGCUCUUCUUCCAGGUGAGUUUCGUA_____UA___________________ .(((((.....)))))((.(((((.((...(((....(((((((((.....))))))))).)))..))))))).))......................................... (-20.12 = -19.57 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:17 2006