
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,777,694 – 6,777,846 |
| Length | 152 |
| Max. P | 0.669248 |

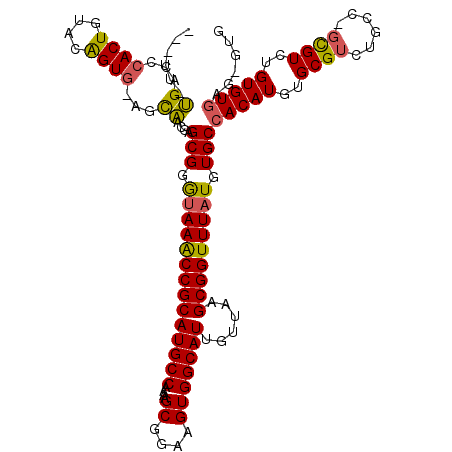
| Location | 6,777,694 – 6,777,814 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -32.18 |
| Energy contribution | -31.97 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6777694 120 - 22224390 CCGUUCUGACCCACUGUACAGUGAAGCAACGAGCGGGUAAACCGCAUGCCAAAAGCGGAAGUGGCAUUGUUAAGCGGUUUAUAUGCCACAUGUGCGUCUGCCAGCGUCUGUGUGAGUGUG ((((((((...((((....))))...))..))))))((((((((((((((....((....)))))))......)))))))))(((((((((..((((......))))..))))).)))). ( -43.40) >DroEre_CAF1 68722 113 - 1 ------UGGCCCACUGUGCAGUGGAGCAACGAGCGGGUAAGCCGCAUGCCAAAAGCGGAAGUGGCAUUGUUAAGCGGUUUAUGUGCCACAUGUGCGUCUGCCUGCGUCUGUGUGAG-GUG ------..((((((.((((((..(((((....(((.((((((((((((((....((....)))))))......))))))))).)))......))).))..).)))).)...))).)-)). ( -44.30) >DroAna_CAF1 83201 115 - 1 AGCUUUCGUACCACUGUCCGGUA-UUUGAGGAGCGGACAAACCGCAUGCCAAAAGCGGAAGUGGCAUUGUUAAGCGGUUUAUGUGCCACAUGUGCGUCUG----UGUCGGUGUAGGGGAG ..((..(.(((.(((((((....-.....)))((((((((((((((((((....((....)))))))......)))))))..(..(.....)..))))))----)..))))))).)..)) ( -39.30) >consensus ___UU_UGACCCACUGUACAGUG_AGCAACGAGCGGGUAAACCGCAUGCCAAAAGCGGAAGUGGCAUUGUUAAGCGGUUUAUGUGCCACAUGUGCGUCUGCC_GCGUCUGUGUGAG_GUG ......((...((((....))))...))....(((.((((((((((((((....((....)))))))......))))))))).)))(((((..((((......))))..)))))...... (-32.18 = -31.97 + -0.21)


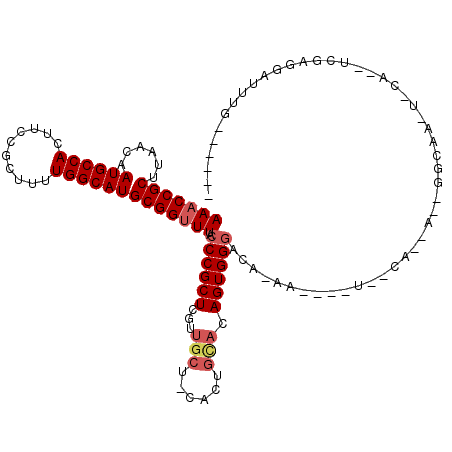
| Location | 6,777,734 – 6,777,846 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.34 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -18.28 |
| Energy contribution | -19.83 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6777734 112 + 22224390 AAACCGCUUAACAAUGCCACUUCCGCUUUUGGCAUGCGGUUUACCCGCUCGUUGCUUCACUGUACAGUGGGUCAGAACGGCUGGCAGCAUUGGCAAAUUCA--UCGAGGAUUUG------ .....(((......(((((...(((.(((((((..((((.....))))........(((((....))))))))))))))).))))).....)))((((((.--....)))))).------ ( -34.20) >DroEre_CAF1 68761 87 + 1 AAACCGCUUAACAAUGCCACUUCCGCUUUUGGCAUGCGGCUUACCCGCUCGUUGCUCCACUGCACAGUGGGCCA-------------------------CA--UCGAGGAUUUG------ ...((........((((((..........))))))((((.....))))..((.((.(((((....))))))).)-------------------------).--....)).....------ ( -24.70) >DroAna_CAF1 83237 119 + 1 AAACCGCUUAACAAUGCCACUUCCGCUUUUGGCAUGCGGUUUGUCCGCUCCUCAAA-UACCGGACAGUGGUACGAAAGCUAUUCCAAAAGGGGCAAUUCCAGGUCAUUAAUUUUCAAUUG ..(((((((.....(((((((((((..(((((...((((.....))))...)))))-...)))).)))))))...))))..((((....))))........)))................ ( -31.90) >consensus AAACCGCUUAACAAUGCCACUUCCGCUUUUGGCAUGCGGUUUACCCGCUCGUUGCU_CACUGCACAGUGGGACA_AA____U__CA__A__GGCAA_U_CA__UCGAGGAUUUG______ (((((((......((((((..........))))))))))))).((((((...(((......))).))))))................................................. (-18.28 = -19.83 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:13 2006