
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,739,556 – 6,739,703 |
| Length | 147 |
| Max. P | 0.997750 |

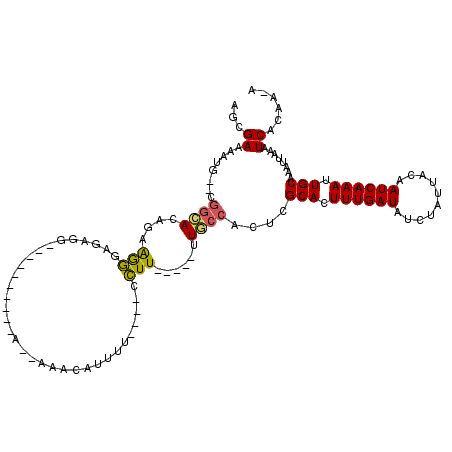
| Location | 6,739,556 – 6,739,665 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -7.82 |
| Energy contribution | -8.58 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6739556 109 - 22224390 A--AAACAUUUU----CCUUUUUUUUUGCCGCUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAAAACCAGAGUUGGGCGAA-CGAAGGAAAAGCGGAAAAAU .--.....((((----((((((((((((.((((((((.((((((..........)))))).)))..............(((....))))))))..-))))))))))).)))))... ( -25.90) >DroSim_CAF1 28478 102 - 1 A--AAACAUUUU----CCUU-----UUGCCGCUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA-ACCAGAGUUGGGUGAA-CGGAGGAAAAGCGGAAAAA- .--.....((((----((((-----((.(((...(((.((((((..........)))))).)))........((((.(-((....)))..)))).-)))....)))).)))))).- ( -21.70) >DroEre_CAF1 29780 104 - 1 A--AAACAUUUU----CCUU-----UUGCCACUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA-GCCAGAGUUGGAUGAAAAGUAGGAAAAGCGGAACAAA .--......(((----((((-----((.(((((.(((.((((((..........)))))).)))........(((...-.((......)).)))..))).))))))).)))).... ( -17.10) >DroAna_CAF1 29974 100 - 1 UACAGAAAUUGUGAACCUUU-----UUCCCACUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA-ACCAGAGUUAAGCGAA-G---GGAAAAGUU------A .................(((-----(((((....(((.((((((..........)))))).)))...((((.((....-....)).)))).....-)---)))))))..------. ( -17.30) >DroPer_CAF1 81840 95 - 1 A--AAAG------AAACCUU-----UUACCACUCGCACUUUGAUAUCUAUUGCAAUCAAAUUGCAAAUUAAAUCACAA-AACAGAGUUAAGCGAU-G----GCAAACCCCAAAA-- (--((((------....)))-----)).(((.((((((((((.......(((((((...))))))).((........)-).))))))...)))))-)----)............-- ( -15.60) >consensus A__AAACAUUUU____CCUU_____UUGCCACUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA_ACCAGAGUUGGGCGAA_CG_AGGAAAAGCGGAAAAA_ ............................(((((.(((.((((((..........)))))).)))...((((.((.........)).))))...............)))))...... ( -7.82 = -8.58 + 0.76)

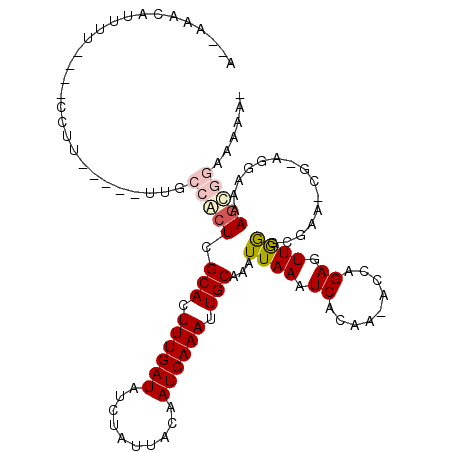

| Location | 6,739,591 – 6,739,703 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -8.57 |
| Energy contribution | -8.21 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6739591 112 - 22224390 AGGGAAAAUG--CGGCACAGAAGGGAGAGAGAGAAAAAAAA--AAACAUUUU----CCUUUUUUUUUGCCGCUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAAAA ...((....(--(((((.(((((((((((.(..........--...).))))----)))))))...))))))..(((.((((((..........)))))).)))........))...... ( -25.92) >DroSim_CAF1 28512 96 - 1 AGGGAAAAUG--CGGCACAGAAGGGAGAGG----------A--AAACAUUUU----CCUU-----UUGCCGCUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA-A ...((....(--(((((..(((((((((..----------.--.....))))----))))-----)))))))..(((.((((((..........)))))).)))........))....-. ( -27.10) >DroEre_CAF1 29816 96 - 1 AGCGAAAGUA--CGGCACAGAAGGGAGAGG----------A--AAACAUUUU----CCUU-----UUGCCACUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA-G .((....)).--.((((..(((((((((..----------.--.....))))----))))-----)))))....(((.((((((..........)))))).)))..............-. ( -24.60) >DroAna_CAF1 30000 100 - 1 AGCGAAAAGG----ACAAAAAAAAAAAAAG----------UACAGAAAUUGUGAACCUUU-----UUCCCACUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA-A ...(((((((----((((............----------........))))...)))))-----)).......(((.((((((..........)))))).)))..............-. ( -11.05) >DroPer_CAF1 81869 94 - 1 --UGAAAUGGAGAGGUACAGAGAGAGCAGA----------A--AAAG------AAACCUU-----UUACCACUCGCACUUUGAUAUCUAUUGCAAUCAAAUUGCAAAUUAAAUCACAA-A --.....((((.((((((((((.(((..(.----------(--((((------....)))-----)).)..)))...))))).))))).(((((((...)))))))......)).)).-. ( -18.40) >consensus AGCGAAAAUG__CGGCACAGAAGGGAGAGG__________A__AAACAUUUU____CCUU_____UUGCCACUCGCACUUUGAUAUCUAUUACAAUCAAAUUGCAAAUUAAAUCACAA_A ...((........((((....(((.................................)))......))))....(((.((((((..........)))))).)))........))...... ( -8.57 = -8.21 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:01 2006