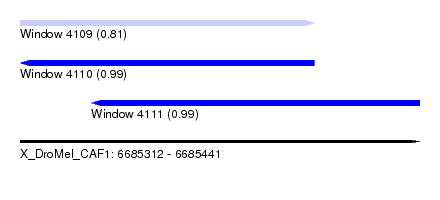
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,685,312 – 6,685,441 |
| Length | 129 |
| Max. P | 0.990408 |

| Location | 6,685,312 – 6,685,407 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.54 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6685312 95 + 22224390 UU----------AACUUUCUU--AAGUUA-AG----AUAUGUUGCUCAACUUACGUGCAAAGUGUGUGGCCACUGGGCCUGAGAUCUACG-----UCCCAACCCAGAGACCAG---ACCC ((----------(((((....--))))))-).----.....((((.(.......).))))...((.(((.(.(((((..((.(((....)-----)).)).))))).).))).---)).. ( -23.00) >DroSec_CAF1 15062 111 + 1 UGAUAUG-AUAUAUGUCUCUUCAGAGUUA-AG----AUAAGUUGCCCAACUUACGUGCAAAGUGUGUGGUCACUGGGCCUGGGAUCUGCGUCGCGUCCCAGCCCAGAGACCAG---ACCC (((...(-((....)))...)))..((..-.(----.((((((....)))))))..)).....((.(((((.((((((.((((((..((...)))))))))))))).))))).---)).. ( -39.20) >DroEre_CAF1 14805 110 + 1 UGAUAUA-ACAUAUCUUCUUU-CAGGUAAGUGACUGAUGUGCUGCCCAACUUACGUGCAAAGUGUUUGGUCACUGGGCCUGGGAUCUACG-----UCCCAACCCAGAGAUUUG---ACCC .(((((.-..)))))..((((-((.((((((.................)))))).)).)))).(((..(((.(((((..((((((....)-----))))).))))).)))..)---)).. ( -32.33) >DroYak_CAF1 14766 109 + 1 UGAAAUAUGCAUAUCU-CUUU-CAAAUUA-UGACUUUAUUUUGGCCCAACUUACGUGCAAAGUAUUUGGUCACUGGGCCUGGGAUCUCCG-----UCCCAACCCAGAGAAUGG---ACCC .............(((-..((-(......-(((((.(((((((..(........)..)))))))...)))))(((((..((((((....)-----))))).))))).))).))---)... ( -27.90) >DroAna_CAF1 15627 95 + 1 -------------GUUUCUAC-CAGCCAAG----------GGCGUCCAACCUACCUCUUGAAGGUUCGAUCUCUGGGCCUGGGGC-ACCAUGGGCCACCAACCCAGAGAUGCGCCAACCA -------------........-....((((----------((.((.......))))))))..(((((((((((((((..((((((-.(....)))).))).))))))))).))..)))). ( -36.60) >consensus UGA_AU____AUAUCUUCUUU_CAAGUUA_AG____AUAUGUUGCCCAACUUACGUGCAAAGUGUUUGGUCACUGGGCCUGGGAUCUACG_____UCCCAACCCAGAGACCAG___ACCC ...................................................................((((.(((((..(((((...........))))).))))).))))......... (-14.40 = -14.64 + 0.24)


| Location | 6,685,312 – 6,685,407 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.54 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.04 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6685312 95 - 22224390 GGGU---CUGGUCUCUGGGUUGGGA-----CGUAGAUCUCAGGCCCAGUGGCCACACACUUUGCACGUAAGUUGAGCAACAUAU----CU-UAACUU--AAGAAAGUU----------AA ..((---.(((((.(((((((((((-----......))))).)))))).))))).))(((((.(...(((((((((........----))-))))))--).)))))).----------.. ( -33.80) >DroSec_CAF1 15062 111 - 1 GGGU---CUGGUCUCUGGGCUGGGACGCGACGCAGAUCCCAGGCCCAGUGACCACACACUUUGCACGUAAGUUGGGCAACUUAU----CU-UAACUCUGAAGAGACAUAUAU-CAUAUCA ..((---((((((.((((((((((((........).))))).)))))).)))).....((((.....((((..((....))...----))-)).....))))))))......-....... ( -40.10) >DroEre_CAF1 14805 110 - 1 GGGU---CAAAUCUCUGGGUUGGGA-----CGUAGAUCCCAGGCCCAGUGACCAAACACUUUGCACGUAAGUUGGGCAGCACAUCAGUCACUUACCUG-AAAGAAGAUAUGU-UAUAUCA .(((---((.....(((((((((((-----......))))).))))))))))).....((((.((.((((((..(((.........))))))))).))-))))..(((((..-.))))). ( -34.30) >DroYak_CAF1 14766 109 - 1 GGGU---CCAUUCUCUGGGUUGGGA-----CGGAGAUCCCAGGCCCAGUGACCAAAUACUUUGCACGUAAGUUGGGCCAAAAUAAAGUCA-UAAUUUG-AAAG-AGAUAUGCAUAUUUCA .(((---(((.((.(((((((((((-----......))))).)))))).)).............((....)))))))).........(((-.....))-)..(-(((((....)))))). ( -33.40) >DroAna_CAF1 15627 95 - 1 UGGUUGGCGCAUCUCUGGGUUGGUGGCCCAUGGU-GCCCCAGGCCCAGAGAUCGAACCUUCAAGAGGUAGGUUGGACGCC----------CUUGGCUG-GUAGAAAC------------- .((((((((..((((((((((((.((((....).-)))))).)))))))))((.(((((((....)).))))).))))))----------...)))).-........------------- ( -40.10) >consensus GGGU___CUAAUCUCUGGGUUGGGA_____CGGAGAUCCCAGGCCCAGUGACCAAACACUUUGCACGUAAGUUGGGCAACAUAU____CA_UAACUUG_AAAGAAGAUAU____AU_UCA ..........(((.(((((((((((...........))))).)))))).)))......(((.((......)).)))............................................ (-18.00 = -17.04 + -0.96)



| Location | 6,685,335 – 6,685,441 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.00 |
| Mean single sequence MFE | -47.70 |
| Consensus MFE | -43.56 |
| Energy contribution | -43.88 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6685335 106 - 22224390 ------ACUGGAACUGGCCAGCUGGGCGCAGUGCGAGGAUGGGUCUGGUCUCUGGGUUGGGA-----CGUAGAUCUCAGGCCCAGUGGCCACACACUUUGCACGUAAGUUGAGCAAC ------..(((......)))(((.(((...((((((((....((.(((((.(((((((((((-----......))))).)))))).))))).)).))))))))....))).)))... ( -46.00) >DroSec_CAF1 15096 111 - 1 ------UCUGGAACUGGCCAGCUGGGCGCAGUGCGAGGAUGGGUCUGGUCUCUGGGCUGGGACGCGACGCAGAUCCCAGGCCCAGUGACCACACACUUUGCACGUAAGUUGGGCAAC ------........((.((((((....((.((((((((....((.(((((.((((((((((((........).))))).)))))).))))).)).)))))))))).)))))).)).. ( -53.00) >DroEre_CAF1 14843 106 - 1 ------ACUGGAACUGGCCAGCUGGACGCAGUGCGAGGAUGGGUCAAAUCUCUGGGUUGGGA-----CGUAGAUCCCAGGCCCAGUGACCAAACACUUUGCACGUAAGUUGGGCAGC ------.......(((.((((((....((.((((((((...(((((.....(((((((((((-----......))))).))))))))))).....)))))))))).)))))).))). ( -47.60) >DroYak_CAF1 14803 112 - 1 GCAGCGACUGGAACUGGCCAGCUGGACGCAGUGCGAGGAUGGGUCCAUUCUCUGGGUUGGGA-----CGGAGAUCCCAGGCCCAGUGACCAAAUACUUUGCACGUAAGUUGGGCCAA ..............(((((((((....((.((((((((...((((......(((((((((((-----......))))).)))))).)))).....)))))))))).)))).))))). ( -44.20) >consensus ______ACUGGAACUGGCCAGCUGGACGCAGUGCGAGGAUGGGUCUAGUCUCUGGGUUGGGA_____CGUAGAUCCCAGGCCCAGUGACCAAACACUUUGCACGUAAGUUGGGCAAC ........(((......)))(((.(((...((((((((....((.(((((.(((((((((((...........))))).)))))).))))).)).))))))))....))).)))... (-43.56 = -43.88 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:40 2006