| Sequence ID | X_DroMel_CAF1 |
|---|---|
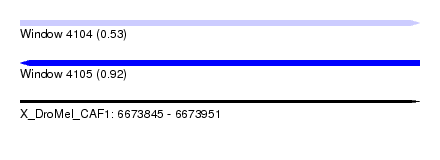
| Location | 6,673,845 – 6,673,951 |
| Length | 106 |
| Max. P | 0.924727 |

| Location | 6,673,845 – 6,673,951 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
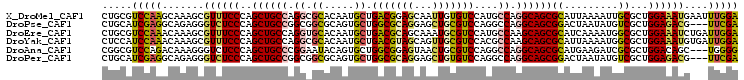
>X_DroMel_CAF1 6673845 106 + 22224390 UCCAAAUUCAUUUCCAGCGCAAUUUUAAUGCGCUGCCUGGCAUGGACACAAUUGCUCCGUCAGCAUUGUGCGCCUGGCAGCUGGGAAACGCUUUGCUUGGACGCAG (((((.....(((((((((((.......)))..((((.(((...(.((((((.(((.....))))))))))))).))))))))))))..((...)))))))..... ( -36.70) >DroPse_CAF1 3765 103 + 1 UCGAA---CGUCUCCAGCGACAUAUUAGUCCGCUGCCUGGCCUGGACGCAGCUCCUGCGCCAGCACUGCGCCGCCGGCAGCUGGGAGACCCUCUGCCUCGAUGCAG .....---.((((((.(.(((......))))((((((.(((..((.(((((....(((....)))))))))))))))))))..))))))...((((......)))) ( -45.00) >DroEre_CAF1 3568 106 + 1 UCCAAUCAGAUUUCCAGCGCCAUUUUGAUGCGCUGCUUGGCAUGGACGCAUUUGCUGCGUCAGCAUUGUGCACCUGGCAGCUGGGAAACGCUUUGUUUGGACGCAG (((((.((((((((((((((((...(((((((((....)))...((((((.....)))))).))))))......)))).))))))))....)))).)))))..... ( -40.90) >DroYak_CAF1 3568 106 + 1 UCCAAUCACAUUUCCAGCGCCAUUUUAAUGCGCUGCUUGGCGUGGACGCAACUGCUACGUCAGCAUUGUGCGCCUGGCAGCUGGGAAACGCUUUGUUUGGAUGGAG (((((.((..((((((((((((.......(((((....)))))((.((((..((((.....))))...)))))))))).))))))))......)).)))))..... ( -39.40) >DroAna_CAF1 3546 103 + 1 CCCCA---GCUGUCCAGCGCGAUCUUCAUGCGCUGCCUGGCCUGGACGCAGUUACUCCGCCAGCACUGUAUUCCGGGCAGCUGGGAGACCCUUUGUCUGGACGCCG .((((---(((((((((((((.......))))))((.((((..(((.(......))))))))))..........)))))))))))((((.....))))........ ( -43.20) >DroPer_CAF1 3776 103 + 1 UCGAA---CGUCUCCAGCGACAUAUUAGUCCGCUGCCUGGCCUGGACACAGCUCCUGCGCCAGCACUGCGCCGCCGGCAGCUGGGAGACCCUCUGCCUCGAUGCAG .....---......(((((((......)).))))).((((((.(((......))).).)))))..(((((.((..(((((..((....))..))))).)).))))) ( -40.20) >consensus UCCAA___CAUUUCCAGCGCCAUUUUAAUGCGCUGCCUGGCCUGGACGCAGCUGCUGCGCCAGCACUGUGCCCCUGGCAGCUGGGAAACCCUUUGCCUGGACGCAG ..............((((((.........)))))).(((((.(((.........))).)))))..((((...((.(((((..((....).).))))).))..)))) (-21.76 = -21.68 + -0.08)



| Location | 6,673,845 – 6,673,951 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -26.15 |
| Energy contribution | -25.35 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6673845 106 - 22224390 CUGCGUCCAAGCAAAGCGUUUCCCAGCUGCCAGGCGCACAAUGCUGACGGAGCAAUUGUGUCCAUGCCAGGCAGCGCAUUAAAAUUGCGCUGGAAAUGAAUUUGGA .....((((((.....(((((((..((((((.(((((((((((((.....))).)))))))....))).))))))(((.......)))...)))))))..)))))) ( -42.40) >DroPse_CAF1 3765 103 - 1 CUGCAUCGAGGCAGAGGGUCUCCCAGCUGCCGGCGGCGCAGUGCUGGCGCAGGAGCUGCGUCCAGGCCAGGCAGCGGACUAAUAUGUCGCUGGAGACG---UUCGA .(((......)))(((.((((((..(((((((((((((((((.(((...)))..))))))))...))).)))))).(((......)))...)))))).---))).. ( -51.00) >DroEre_CAF1 3568 106 - 1 CUGCGUCCAAACAAAGCGUUUCCCAGCUGCCAGGUGCACAAUGCUGACGCAGCAAAUGCGUCCAUGCCAAGCAGCGCAUCAAAAUGGCGCUGGAAAUCUGAUUGGA .....(((((....((.((((((.(((.(((((((((....((((((((((.....)))))).......))))..)))))....)))))))))))))))..))))) ( -37.21) >DroYak_CAF1 3568 106 - 1 CUCCAUCCAAACAAAGCGUUUCCCAGCUGCCAGGCGCACAAUGCUGACGUAGCAGUUGCGUCCACGCCAAGCAGCGCAUUAAAAUGGCGCUGGAAAUGUGAUUGGA .....(((((.....((((((((.(((.((((((((((...(((((...)))))..))))))..(((......)))........)))))))))))))))..))))) ( -39.50) >DroAna_CAF1 3546 103 - 1 CGGCGUCCAGACAAAGGGUCUCCCAGCUGCCCGGAAUACAGUGCUGGCGGAGUAACUGCGUCCAGGCCAGGCAGCGCAUGAAGAUCGCGCUGGACAGC---UGGGG ((((((((((.....(((((((...((((((.((.........((((((.((...)).)).)))).)).))))))....).))))).).)))))).))---))... ( -40.70) >DroPer_CAF1 3776 103 - 1 CUGCAUCGAGGCAGAGGGUCUCCCAGCUGCCGGCGGCGCAGUGCUGGCGCAGGAGCUGUGUCCAGGCCAGGCAGCGGACUAAUAUGUCGCUGGAGACG---UUCGA .(((......)))(((.((((((..(((((((((((((((((.(((...)))..))))))))...))).)))))).(((......)))...)))))).---))).. ( -49.10) >consensus CUGCAUCCAAACAAAGCGUCUCCCAGCUGCCAGGCGCACAAUGCUGACGCAGCAACUGCGUCCAGGCCAGGCAGCGCAUUAAAAUGGCGCUGGAAAUG___UUGGA .....(((((.......((((((..((((((.((.((.....)).(((((((...)))))))....)).))))))((.........))...))))))....))))) (-26.15 = -25.35 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:35 2006