
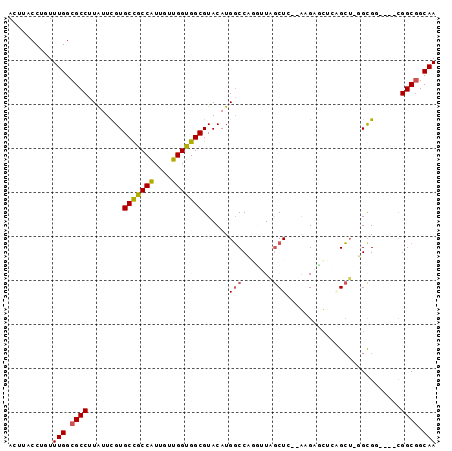
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,665,912 – 6,666,003 |
| Length | 91 |
| Max. P | 0.597576 |

| Location | 6,665,912 – 6,666,003 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -22.55 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6665912 91 + 22224390 ACUUACCUGUUUGGCGCCUUGUUCGUGCCGCCAUUGUUGGUGGCGUACAUGGCCCGGUUGGCUG--GAGACCUUAGCUCGGCGU----UGGCGGCAA ......(((.(..(((((..(((...(((((((....)))))))......(((((((....)))--).).))..)))..)))))----..))))... ( -35.40) >DroVir_CAF1 18060 90 + 1 ACUCACCUAUUUGGCGCCUUAUUUGUGCUGCCAUUAUUGGUGGCGUACAUGGCCACGUUAGCUG--AGGUGU--GGC---GUGUUGGCCGGCUGCAA .............(((((.....(((((.(((((.....)))))))))).((((((((((((..--..)).)--)))---))...))))))).)).. ( -31.30) >DroPse_CAF1 17525 91 + 1 ACGUACCUGUUUGGCGCCUUAUUCGUGCCGCCAUUGUUGGUGGCGUACAUGGCCAAUGUAGCUC--ACGUGCUCUGCUAUGCGG----CGGCGGCAA ..........(((.((((........(((((((....)))))))((.((((((....(.(((..--....)))).))))))..)----))))).))) ( -33.90) >DroSec_CAF1 14705 91 + 1 ACUUACCUGUUUGGCGCCUUGUUCGUGCCGCCAUUGUUGGUGGCGUACAUGGCCCGGAUGGCUG--GAGACCUUAGCUCGGUGU----UGGCGGCAA .....((.((((((.(((.(((...((((((((....)))))))).))).))))))))).((..--(..(((.......))).)----..))))... ( -35.60) >DroAna_CAF1 17747 92 + 1 AUUUACCUGUUUGGCGCCUUGUUUGUGCCACCGUUAUUGGUGGCGUACAUGUCCCGGUUGACUCACGUGAUCUGAGGU-GGCGG----UGGCGGCAA .....((.....)).(((.(((...((((((((....)))))))).))).((((((.(...((((.......))))..-).)))----.)))))).. ( -31.60) >DroPer_CAF1 17841 91 + 1 ACGUACCUGUUUGGCGCCUUAUUCGUGCCGCCAUUGUUGGUGGCGUACAUGGCCAAUGUAGCUC--ACGUGUUCUGCUAUGCGG----CGGCGGCAA ..........(((.((((........(((((((....))))))).......(((...(((((..--.........)))))..))----))))).))) ( -33.00) >consensus ACUUACCUGUUUGGCGCCUUAUUCGUGCCGCCAUUGUUGGUGGCGUACAUGGCCAGGUUAGCUC__AAGAGCUCAGCU_GGCGG____CGGCGGCAA ..........(((.((((........(((((((....)))))))......(((.......)))..........................)))).))) (-22.55 = -22.63 + 0.08)


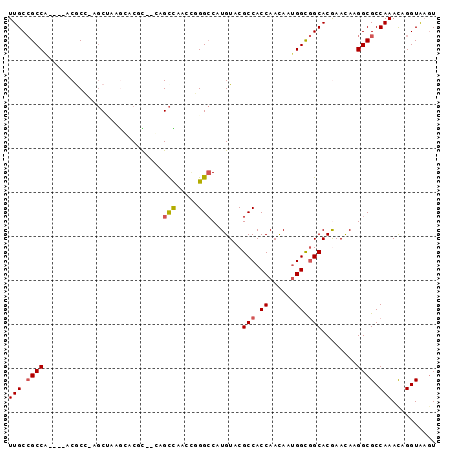
| Location | 6,665,912 – 6,666,003 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.93 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6665912 91 - 22224390 UUGCCGCCA----ACGCCGAGCUAAGGUCUC--CAGCCAACCGGGCCAUGUACGCCACCAACAAUGGCGGCACGAACAAGGCGCCAAACAGGUAAGU (((((((..----..(((.......)))...--..)).....(((((.(((..(((.(((....))).)))....))).))).)).....))))).. ( -26.50) >DroVir_CAF1 18060 90 - 1 UUGCAGCCGGCCAACAC---GCC--ACACCU--CAGCUAACGUGGCCAUGUACGCCACCAAUAAUGGCAGCACAAAUAAGGCGCCAAAUAGGUGAGU ........(((......---)))--....((--((.(((..(((((.......)))))......((((.((.(......)))))))..))).)))). ( -26.20) >DroPse_CAF1 17525 91 - 1 UUGCCGCCG----CCGCAUAGCAGAGCACGU--GAGCUACAUUGGCCAUGUACGCCACCAACAAUGGCGGCACGAAUAAGGCGCCAAACAGGUACGU ..(((((((----((((........))..((--(.(((((((.....))))).))))).......))))))........)))(((.....))).... ( -27.90) >DroSec_CAF1 14705 91 - 1 UUGCCGCCA----ACACCGAGCUAAGGUCUC--CAGCCAUCCGGGCCAUGUACGCCACCAACAAUGGCGGCACGAACAAGGCGCCAAACAGGUAAGU (((((((..----..(((.......)))...--..)).....(((((.(((..(((.(((....))).)))....))).))).)).....))))).. ( -26.20) >DroAna_CAF1 17747 92 - 1 UUGCCGCCA----CCGCC-ACCUCAGAUCACGUGAGUCAACCGGGACAUGUACGCCACCAAUAACGGUGGCACAAACAAGGCGCCAAACAGGUAAAU (((((....----.((((-.(((......((....)).....)))........((((((......))))))........)))).......))))).. ( -28.50) >DroPer_CAF1 17841 91 - 1 UUGCCGCCG----CCGCAUAGCAGAACACGU--GAGCUACAUUGGCCAUGUACGCCACCAACAAUGGCGGCACGAAUAAGGCGCCAAACAGGUACGU .((((((((----(......)).......((--(.(((((((.....))))).))))).......)))))))........(..((.....))..).. ( -26.80) >consensus UUGCCGCCA____ACGCC_AGCUAAGCACGC__CAGCCAACCGGGCCAUGUACGCCACCAACAAUGGCGGCACGAACAAGGCGCCAAACAGGUAAGU (((.((((...........................(((.....))).......(((.((......)).)))........)))).))).......... (-17.82 = -17.93 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:28 2006