
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,653,399 – 6,653,500 |
| Length | 101 |
| Max. P | 0.611337 |

| Location | 6,653,399 – 6,653,500 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -37.31 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.54 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
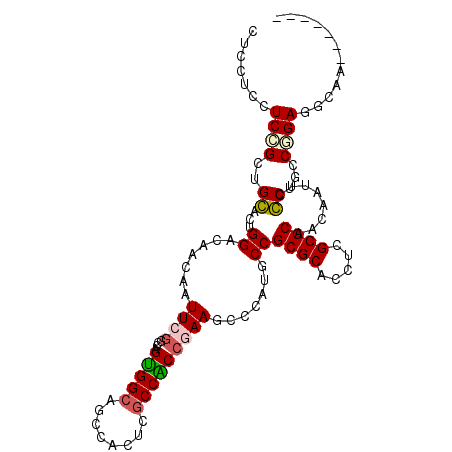
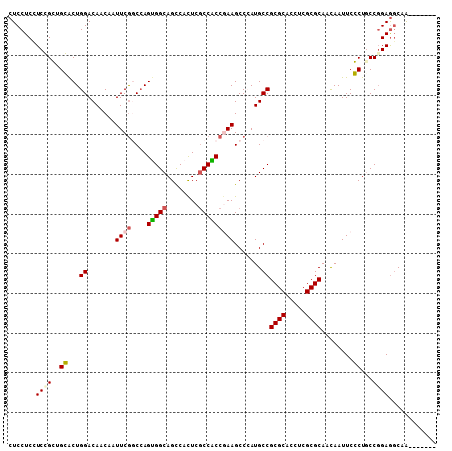
>X_DroMel_CAF1 6653399 101 + 22224390 CUCCUCCUCCGGUGGACUGGACAACCAUUCGGCCAGUGGCAGCCACUCGCCACCGAAGCCCAUGCCGCGCACAUCGCGCAGCAAUUCCCUUCCGGAGGCCA------- .....((((((((.((.(((....))).))(((.(((((...))))).))))))((((....(((.((((.....)))).))).....)))).)))))...------- ( -40.40) >DroVir_CAF1 2551 108 + 1 CUCCUCCUCGGCGGCACUGGACACCAAUUCGCCCAGCGGCAGCCAUUCGCCGCCAAAGCCAAUGCCGCGCACCUCGCGCAACAAUUCGCUGCCCGAAGCGGUCAAUCC ..((.(.((((((((...((......(((.((...(((((........)))))....)).))).))((((.....))))........)))).)))).).))....... ( -33.80) >DroGri_CAF1 2971 107 + 1 CUCCUCCUCGGCGGCGCUGGACACCAAUUCGGCCAGUGGCAGUCACUCGCCGCCCAAGCCCAUGCCGCGCACUUCGCGCAAUAAUUCGCUGCCGGAGGCAACAA-GCU ..(((((..(((((((((((..........(((.((((.....)))).)))((....))))).)).((((.....))))........)))))))))))......-... ( -41.40) >DroSec_CAF1 2211 101 + 1 CUCCUCCUCCGGUGGACUGGACAACCAUUCAGCCAGUGGCAGCCACUCGCCACCGAAGCCCAUGCCGCGCACCUCGCGCAACAAUUCCCUGCCGGAGGCCA------- .....(((((((..((((((.(.........))))))((((.....(((....)))......))))((((.....)))).........)..)))))))...------- ( -37.20) >DroEre_CAF1 2196 101 + 1 CUCCUCGUCCGGUGGACUGGACAACCAUUCGGCCAGUGGCAGCCACUCCCCACCGAAGCCCAUGCCGCGCACUUCGCGCAACAAUUCCCUACCGGAGGCUA------- .....(.((((((((...((.......(((((..(((((...))))).....))))).......))((((.....)))).........)))))))).)...------- ( -34.34) >DroMoj_CAF1 2361 108 + 1 CUCCUCAUCGGCUGCACUGGACACCAAUUCGCCAAGCGGCAGCCACUCGCCGCCGAAGCCGAUGCCGCGCACCUCGCGCAACAAUUCGCUGCCGGAGUCAACAACGCC ((((.((((((((.(..(((.(........)))).(((((........))))).).))))))))..((((.....))))..............))))........... ( -36.70) >consensus CUCCUCCUCCGCUGCACUGGACAACAAUUCGGCCAGUGGCAGCCACUCGCCACCGAAGCCCAUGCCGCGCACCUCGCGCAACAAUUCCCUGCCGGAGGCAA_______ .......((((..((...((.......((((....(((((........))))))))).......))((((.....))))........))...))))............ (-22.68 = -22.54 + -0.14)

| Location | 6,653,399 – 6,653,500 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -48.52 |
| Consensus MFE | -33.87 |
| Energy contribution | -34.23 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
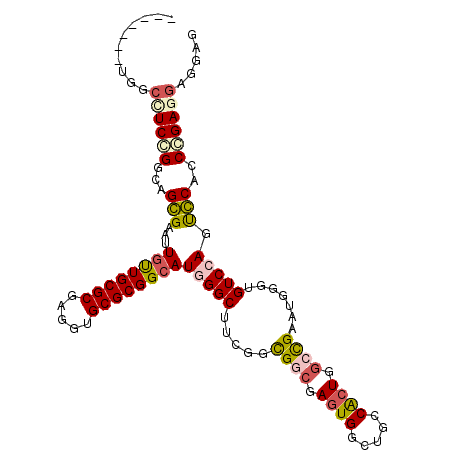
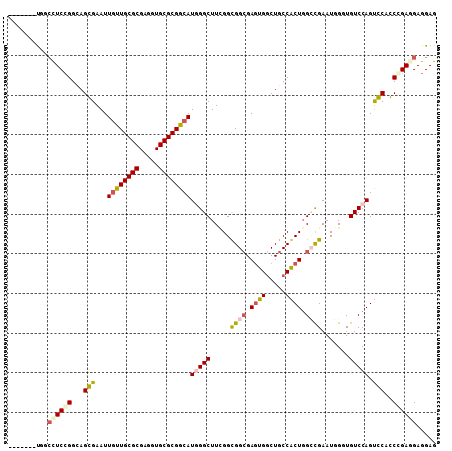
>X_DroMel_CAF1 6653399 101 - 22224390 -------UGGCCUCCGGAAGGGAAUUGCUGCGCGAUGUGCGCGGCAUGGGCUUCGGUGGCGAGUGGCUGCCACUGGCCGAAUGGUUGUCCAGUCCACCGGAGGAGGAG -------...(((((((...(((..((((((((.....))))))))(((((.(((.((((.(((((...))))).))))..)))..))))).))).)))))))..... ( -52.90) >DroVir_CAF1 2551 108 - 1 GGAUUGACCGCUUCGGGCAGCGAAUUGUUGCGCGAGGUGCGCGGCAUUGGCUUUGGCGGCGAAUGGCUGCCGCUGGGCGAAUUGGUGUCCAGUGCCGCCGAGGAGGAG .......((((((((.(((((.....))))).))))))(.(((((.........((((((.....))))))((((((((......))))))))))))))..))..... ( -51.40) >DroGri_CAF1 2971 107 - 1 AGC-UUGUUGCCUCCGGCAGCGAAUUAUUGCGCGAAGUGCGCGGCAUGGGCUUGGGCGGCGAGUGACUGCCACUGGCCGAAUUGGUGUCCAGCGCCGCCGAGGAGGAG ...-......((((((((...........((((.....))))(((.((((((..(.((((.((((.....)))).))))..)..).)))))..))))))..))))).. ( -47.90) >DroSec_CAF1 2211 101 - 1 -------UGGCCUCCGGCAGGGAAUUGUUGCGCGAGGUGCGCGGCAUGGGCUUCGGUGGCGAGUGGCUGCCACUGGCUGAAUGGUUGUCCAGUCCACCGGAGGAGGAG -------...(((((((...(((..((((((((.....))))))))(((((.(((..(((.(((((...))))).)))...)))..))))).))).)))))))..... ( -48.40) >DroEre_CAF1 2196 101 - 1 -------UAGCCUCCGGUAGGGAAUUGUUGCGCGAAGUGCGCGGCAUGGGCUUCGGUGGGGAGUGGCUGCCACUGGCCGAAUGGUUGUCCAGUCCACCGGACGAGGAG -------...((((((((.(((...((((((((.....))))))))(((((((((((.((..((....))..)).)))))).....))))).)))))))))...)).. ( -42.80) >DroMoj_CAF1 2361 108 - 1 GGCGUUGUUGACUCCGGCAGCGAAUUGUUGCGCGAGGUGCGCGGCAUCGGCUUCGGCGGCGAGUGGCUGCCGCUUGGCGAAUUGGUGUCCAGUGCAGCCGAUGAGGAG ..((((((((....))))))))....(((((((.....)))))))(((((((..((((((.....))))))((((((((......)).)))).)))))))))...... ( -47.70) >consensus _______UGGCCUCCGGCAGCGAAUUGUUGCGCGAGGUGCGCGGCAUGGGCUUCGGCGGCGAGUGGCUGCCACUGGCCGAAUGGGUGUCCAGUCCACCCGAGGAGGAG ..........((((((...(((...((((((((.....))))))))(((((.....((((.((((.....)))).)))).......))))).)))..))))))..... (-33.87 = -34.23 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:25 2006