| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,640,343 – 6,640,447 |
| Length | 104 |
| Max. P | 0.826374 |

| Location | 6,640,343 – 6,640,447 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6640343 104 + 22224390 CCCUGCCAAUGAUGACGUCACUCGCAGGUGCUGUCAUCACAAAAGUAUGUGCAUUUAUUUUCGU----UGUGUACGAUUUGUGCGAUUUGUACGAUUUGUACGAUUUU ...(((((..(((((((.((((....)))).)))))))((....)).)).)))...........----..(((((((.((((((.....)))))).)))))))..... ( -28.10) >DroSec_CAF1 34173 107 + 1 CCCUGACAAUGAUGACGUCACUCGCAGGUGCUGUCAUCACAAAAGUAUGUGCAUUUGUUUUCGUACGAUUUGUACGAUUUGUACGAUUUGUACGAUUUGUACGAUU-U .........((((((((.((((....)))).))))))))........((((((.......((((((((.((((((.....)))))).))))))))..))))))...-. ( -34.70) >DroSim_CAF1 31949 89 + 1 CCCUGACAAUGAUGACGUCACUCGCAGGUGCUGUCAUCACAAAAGUAUGUGCAUUUGUUUUCGUACGAUUUGUACG------------------AUUUGUACGAUU-U .........((((((((.((((....)))).))))))))........((((((.......((((((.....)))))------------------)..))))))...-. ( -24.80) >DroEre_CAF1 27858 85 + 1 CCCUGCCAAUGAUGACGUCACUCGCAGGUGCUGUCAUCAGAAAAGUAUGUGCAUUUGUUUUCGC----UGUGUACG------------------AUUUGUGGGAUU-U .((..(...((((((((.((((....)))).))))))))...((((.(((((((..((....))----.)))))))------------------)))))..))...-. ( -26.00) >DroYak_CAF1 30182 93 + 1 CCCAGGCAACGAUGACGUCACUCGCAGGUGCUGUCAUCGCAAAAGUAUGUGCAUUUGUUUUCGC----UGUGUACGAU----------UGUGCGAUUUGUGCGAUU-U ....(....)(((((((.((((....)))).)))))))(((.((((.((..((.((((......----.....)))).----------))..)))))).)))....-. ( -27.20) >consensus CCCUGACAAUGAUGACGUCACUCGCAGGUGCUGUCAUCACAAAAGUAUGUGCAUUUGUUUUCGU____UGUGUACGAU__________UGU_CGAUUUGUACGAUU_U .........((((((((.((((....)))).)))))))).........((((((...............))))))................................. (-17.94 = -18.02 + 0.08)

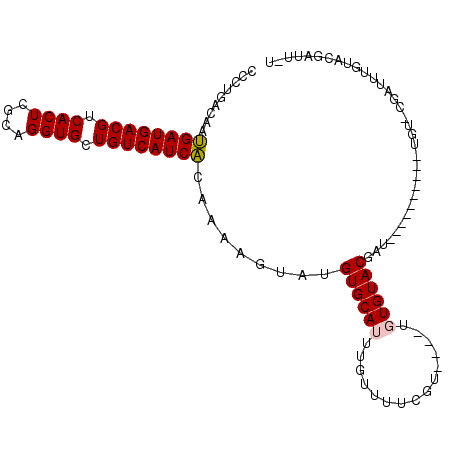

| Location | 6,640,343 – 6,640,447 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6640343 104 - 22224390 AAAAUCGUACAAAUCGUACAAAUCGCACAAAUCGUACACA----ACGAAAAUAAAUGCACAUACUUUUGUGAUGACAGCACCUGCGAGUGACGUCAUCAUUGGCAGGG ......((((.....)))).....((.....((((.....----)))).........((((......))))......)).(((((.(((((.....))))).))))). ( -26.90) >DroSec_CAF1 34173 107 - 1 A-AAUCGUACAAAUCGUACAAAUCGUACAAAUCGUACAAAUCGUACGAAAACAAAUGCACAUACUUUUGUGAUGACAGCACCUGCGAGUGACGUCAUCAUUGUCAGGG .-....(((......((((.....))))...((((((.....)))))).......))).....(((..((((((((..(((......)))..))))))))....))). ( -25.70) >DroSim_CAF1 31949 89 - 1 A-AAUCGUACAAAU------------------CGUACAAAUCGUACGAAAACAAAUGCACAUACUUUUGUGAUGACAGCACCUGCGAGUGACGUCAUCAUUGUCAGGG .-....(.((((.(------------------(((((.....))))))..(((((..........)))))((((((..(((......)))..)))))).))))).... ( -20.90) >DroEre_CAF1 27858 85 - 1 A-AAUCCCACAAAU------------------CGUACACA----GCGAAAACAAAUGCACAUACUUUUCUGAUGACAGCACCUGCGAGUGACGUCAUCAUUGGCAGGG .-...(((.....(------------------(((.....----)))).......(((.((........(((((((..(((......)))..))))))).)))))))) ( -20.10) >DroYak_CAF1 30182 93 - 1 A-AAUCGCACAAAUCGCACA----------AUCGUACACA----GCGAAAACAAAUGCACAUACUUUUGCGAUGACAGCACCUGCGAGUGACGUCAUCGUUGCCUGGG .-....(((......(((..----------.((((.....----)))).......)))..........((((((((..(((......)))..)))))))))))..... ( -20.30) >consensus A_AAUCGUACAAAUCG_ACA__________AUCGUACACA____ACGAAAACAAAUGCACAUACUUUUGUGAUGACAGCACCUGCGAGUGACGUCAUCAUUGGCAGGG ...............................................................(((..((((((((..(((......)))..))))))))....))). (-13.62 = -13.70 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:21 2006