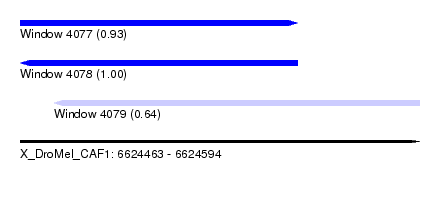
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,624,463 – 6,624,594 |
| Length | 131 |
| Max. P | 0.999694 |

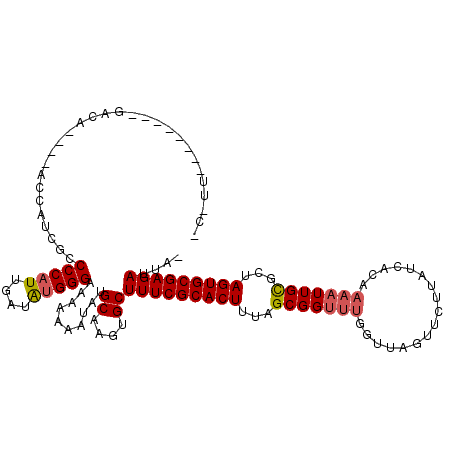
| Location | 6,624,463 – 6,624,554 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.49 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -20.97 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6624463 91 + 22224390 -----------------------------CCCAUUGAUAUGGGUAAAAAAUAUGCAAGUGCUUUCGCACUUCAGCGGUUUAGUUAGUUCUUAUCACAAAAUUGCGCUAGUGCGAGAUUAA -----------------------------(((((....)))))..........((....))(((((((((...(((((((.((...........)).)))))))...))))))))).... ( -25.30) >DroSim_CAF1 16214 119 + 1 UCGUUUCAUUUUGGACAGAAGACCAUCGCCCCAUUGAUAGGGGAAUAAAAUAUGCAAGUGCUUUCGCACUUUAGCGGUUUGGUUAGUUCUUAUCACAAAAUUGCGCUAGUGCGAGAUUA- ..(((((((((((((.(((((((((...((((.......)))).........(((((((((....))))))..)))...)))))..))))..)).)))))).(((....))))))))..- ( -30.70) >DroEre_CAF1 12521 103 + 1 ACUUUG-------GACA---CACCAUCGCCCCAUUGAUGUGGGA-ACAAAUAUGCAAGUGCUUUCGCACUUUGGCGGUUGGGUUAGUUCUUAUCA-----UUGUGCUAGUGCGAGAUUA- ..(((.-------(.((---(.((((((((((((....))))..-..........((((((....)))))).))))).)))..((((.(......-----..).)))))))).)))...- ( -27.60) >consensus _C_UU________GACA____ACCAUCGCCCCAUUGAUAUGGGAAAAAAAUAUGCAAGUGCUUUCGCACUUUAGCGGUUUGGUUAGUUCUUAUCACAAAAUUGCGCUAGUGCGAGAUUA_ .............................(((((....)))))..........((....))(((((((((...(((((((.................)))))))...))))))))).... (-20.97 = -21.86 + 0.89)


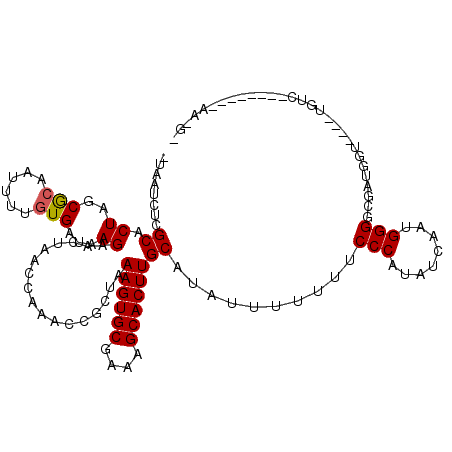
| Location | 6,624,463 – 6,624,554 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.49 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6624463 91 - 22224390 UUAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACUAAACCGCUGAAGUGCGAAAGCACUUGCAUAUUUUUUACCCAUAUCAAUGGG----------------------------- ........((..(((((.......((.......)).........)))))((((((....))))))))..........(((((....)))))----------------------------- ( -22.29) >DroSim_CAF1 16214 119 - 1 -UAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCCCUAUCAAUGGGGCGAUGGUCUUCUGUCCAAAAUGAAACGA -......(((....)))..((((((.((((.(((...((((....((..((((((....)))))))).........(((((((....))))).)))))).)))))))))))))....... ( -33.00) >DroEre_CAF1 12521 103 - 1 -UAAUCUCGCACUAGCACAA-----UGAUAAGAACUAACCCAACCGCCAAAGUGCGAAAGCACUUGCAUAUUUGU-UCCCACAUCAAUGGGGCGAUGGUG---UGUC-------CAAAGU -......(((((((......-----....................((..((((((....))))))))....((((-.((((......)))))))))))))---))..-------...... ( -27.00) >consensus _UAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUUUUCCCAUAUCAAUGGGGCGAUGGU____UGUC________AA_G_ ........((.((..(((......)))...)).................((((((....))))))))..........(((........)))............................. (-17.69 = -17.80 + 0.11)


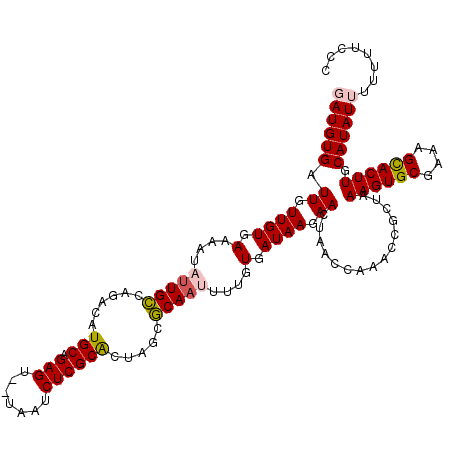
| Location | 6,624,474 – 6,624,594 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.76 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6624474 120 - 22224390 UAUGUGAUUGUUGUGAAAAUAUUGCUAGACAUGCAGAGUUUUAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACUAAACCGCUGAAGUGCGAAAGCACUUGCAUAUUUUUUACCC ...((..((((((..(((....((((((...(((.(((.......))))))))))))...)))..))))))..))..........((..((((((....))))))))............. ( -30.60) >DroSec_CAF1 18429 117 - 1 GAUGUGAUUGUUGUGAAAAUAUUGCCAGGCGUGCAGAGU--UAAUCUCGCGCUAGCGCA-UUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCC ...((..((((((..(((....((((.((((((.(((..--...))))))))).).)))-)))..))))))..))..........((..((((((....))))))))............. ( -33.60) >DroSim_CAF1 16254 118 - 1 GAUGUGAUUGUUGUGAAAAUAUUGCCAGGCGUGCAGAGU--UAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUAUUCCC ...((..((((((..((...(((((...((((((.(((.--....)))))))..)))))))))..))))))..))..........((..((((((....))))))))............. ( -30.40) >DroEre_CAF1 12551 112 - 1 UAUGUGAUUGUUGUGAAAAUAUUGUCAGACAUGCAGAGU--UAAUCUCGCACUAGCACAA-----UGAUAAGAACUAACCCAACCGCCAAAGUGCGAAAGCACUUGCAUAUUUGU-UCCC (((((.((..(((((.......((.....))(((.(((.--....))))))....)))))-----..))....................((((((....))))))))))).....-.... ( -25.40) >DroYak_CAF1 12816 112 - 1 GAUGUGAUUGUUGUGAAAACAUUGCCAGACAUGCAGAGU--UAAUCUCGCAC-AGCGCAA-----UGAUAAGAACUAAACAAACCGCUAAAGUGCGAAAGUACUUGCAUAUUUUUGUACC ((((((.(((((((.....((((((......(((.(((.--....)))))).-...))))-----))......))..))))).......((((((....)))))).))))))........ ( -24.80) >consensus GAUGUGAUUGUUGUGAAAAUAUUGCCAGACAUGCAGAGU__UAAUCUCGCACUAGCGCAAUUUUGUGAUAAGAACUAACCAAACCGCUAAAGUGCGAAAGCACUUGCAUAUUUUUUUCCC ((((((.((.((((.(....(((((......(((.(((.......)))))).....)))))....).)))).))...............((((((....)))))).))))))........ (-22.12 = -22.48 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:12 2006