
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 838,966 – 839,081 |
| Length | 115 |
| Max. P | 0.999238 |

| Location | 838,966 – 839,081 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.25 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -15.88 |
| Energy contribution | -17.05 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
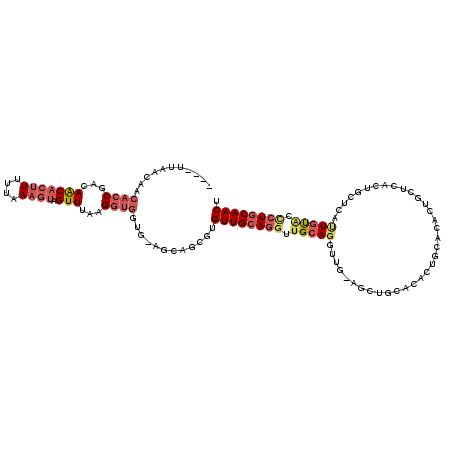
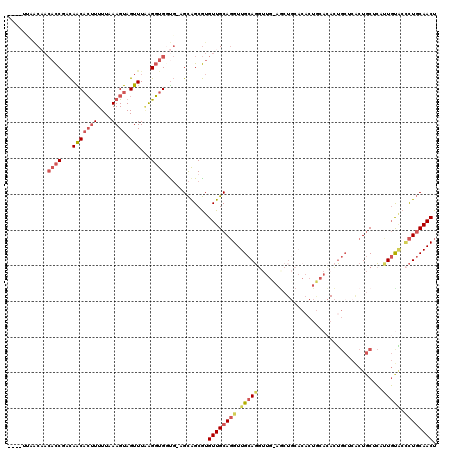
>X_DroMel_CAF1 838966 115 + 22224390 ----UCAACAACACCGACAACACUUUUUCAAGUAGUUUAGGGUGGUG-AGCAGCGUGUUGCAGGUUGCAGGUUGUAGCUGCACACUGCACACUGCUCACUGCUCAUUGUACCCUGCAACU ----.................((((....))))((((((((((((((-(((((.(....((((..(((((..(((....)))..)))))..)))).).))))))))..))))))).)))) ( -41.00) >DroSec_CAF1 39829 115 + 1 ----UUAACAACACCGACAACACUUUUUCAAGUAGUUUGAGGUGGUG-AGCAGCGUGUUGCAGGUUGCAGGUUGCAGCUGCACACUGCACACUGCUCACUGCUCUUUGUACCCUGCAACU ----...((((((((...(((((((....)))).)))...))))(((-(((((.(((((((((.((((.....)))))))))....)))).))))))))......))))........... ( -41.10) >DroSim_CAF1 36076 115 + 1 ----UUAACAACACCGACAACACUUUUUCAAGUAGUUUGAGGUGGUG-AGCAGCGUGUUGCAGGUUGCAGGUUGCAGCUGCACACUGCACACUGCUCACUGCUCUUUGUACCCUGCAACU ----...((((((((...(((((((....)))).)))...))))(((-(((((.(((((((((.((((.....)))))))))....)))).))))))))......))))........... ( -41.10) >DroEre_CAF1 39336 99 + 1 ACCAUCCUCCACACCAACAGCACUUUUUAAAGUAGUUUAAGGUGCCG-AGCAGCGUGUUGCAGGUUGCAGGU--------------------UGCACACUGCACACUGCAGCCUGCAACU ......(((..((((...(((((((....)))).)))...))))..)-))......((((((((((((((..--------------------(((.....)))..)))))))))))))). ( -41.10) >DroYak_CAF1 36705 112 + 1 -------GCAACACCAACAGCUCCUUUUAAAGUAGUUUAAGGUGCUG-AGCAGCGUGUUGCAGGUUGCAGCUGCACACUGAACACUAAACACUGCACACUGCACACUGUGCUCUGCAACU -------((((((((..((((.((((............)))).))))-....).))))))).((((((((..(((((.((............(((.....))))).))))).)))))))) ( -38.90) >DroAna_CAF1 38785 95 + 1 ----UUAAC----UCUUUAACAAUUUUUAAAGUAGUUUUAGUUGGUUCUGUGGCAUGUUGCAUGUUGCAUG-----GCUCC--------UAUG----GCUCCUAUUUUCUAGUUUCAACU ----..(((----(((((((......)))))).))))..((((((..(((..(((((...)))))..)..(-----(((..--------...)----)))..........))..)))))) ( -20.40) >consensus ____UUAACAACACCGACAACACUUUUUAAAGUAGUUUAAGGUGGUG_AGCAGCGUGUUGCAGGUUGCAGGUUG_AGCUGCACACUGCACACUGCUCACUGCUCAUUGUACCCUGCAACU ...........((((...(((((((....)))).)))...))))............((((((((.(((((...................................))))).)))))))). (-15.88 = -17.05 + 1.17)

| Location | 838,966 – 839,081 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.25 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -14.98 |
| Energy contribution | -18.12 |
| Covariance contribution | 3.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.38 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
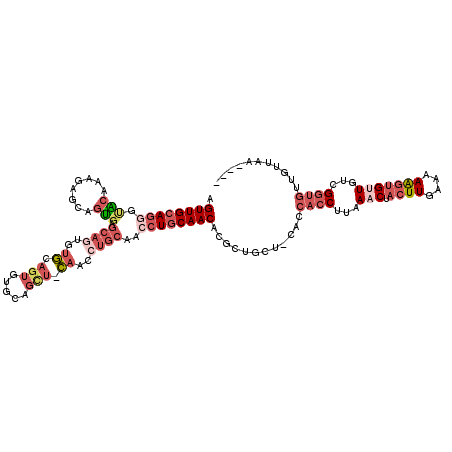
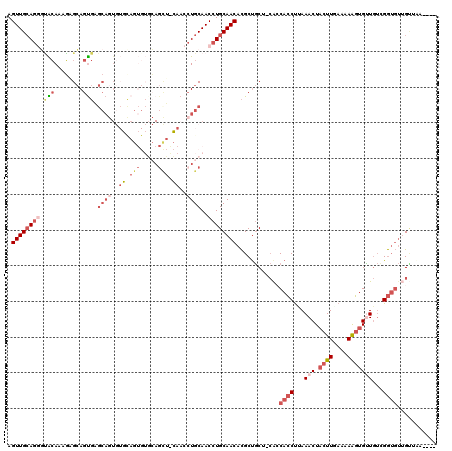
>X_DroMel_CAF1 838966 115 - 22224390 AGUUGCAGGGUACAAUGAGCAGUGAGCAGUGUGCAGUGUGCAGCUACAACCUGCAACCUGCAACACGCUGCU-CACCACCCUAAACUACUUGAAAAAGUGUUGUCGGUGUUGUUGA---- ............(((..(...(((((((((((((((..(((((.......)))))..))))...))))))))-)))((((...(((.((((....)))))))...)))))..))).---- ( -45.00) >DroSec_CAF1 39829 115 - 1 AGUUGCAGGGUACAAAGAGCAGUGAGCAGUGUGCAGUGUGCAGCUGCAACCUGCAACCUGCAACACGCUGCU-CACCACCUCAAACUACUUGAAAAAGUGUUGUCGGUGUUGUUAA---- ...........((((.((((((((.((((..(((((..(((....)))..)))))..))))....)))))))-)..((((...(((.((((....)))))))...))))))))...---- ( -43.90) >DroSim_CAF1 36076 115 - 1 AGUUGCAGGGUACAAAGAGCAGUGAGCAGUGUGCAGUGUGCAGCUGCAACCUGCAACCUGCAACACGCUGCU-CACCACCUCAAACUACUUGAAAAAGUGUUGUCGGUGUUGUUAA---- ...........((((.((((((((.((((..(((((..(((....)))..)))))..))))....)))))))-)..((((...(((.((((....)))))))...))))))))...---- ( -43.90) >DroEre_CAF1 39336 99 - 1 AGUUGCAGGCUGCAGUGUGCAGUGUGCA--------------------ACCUGCAACCUGCAACACGCUGCU-CGGCACCUUAAACUACUUUAAAAAGUGCUGUUGGUGUGGAGGAUGGU .((((((((.(((((..(((.....)))--------------------..))))).)))))))).((.(.((-(.(((((......(((((....))))).....))))).))).))).. ( -42.80) >DroYak_CAF1 36705 112 - 1 AGUUGCAGAGCACAGUGUGCAGUGUGCAGUGUUUAGUGUUCAGUGUGCAGCUGCAACCUGCAACACGCUGCU-CAGCACCUUAAACUACUUUAAAAGGAGCUGUUGGUGUUGC------- .(((((((.(((((.((.(((.((.((...)).)).))).)).)))))..)))))))..(((((((.(....-((((.((((............)))).))))..))))))))------- ( -42.10) >DroAna_CAF1 38785 95 - 1 AGUUGAAACUAGAAAAUAGGAGC----CAUA--------GGAGC-----CAUGCAACAUGCAACAUGCCACAGAACCAACUAAAACUACUUUAAAAAUUGUUAAAGA----GUUAA---- (((((.............((..(----(...--------))..)-----).(((.....)))..............)))))..((((.((((((......)))))))----)))..---- ( -16.00) >consensus AGUUGCAGGGUACAAAGAGCAGUGAGCAGUGUGCAGUGUGCAGCU_CAACCUGCAACCUGCAACACGCUGCU_CACCACCUUAAACUACUUGAAAAAGUGUUGUCGGUGUUGUUAA____ .((((((((.(((........))).((((..((.(((.....))).))..))))..))))))))............((((...(((.((((....)))))))...))))........... (-14.98 = -18.12 + 3.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:57 2006