
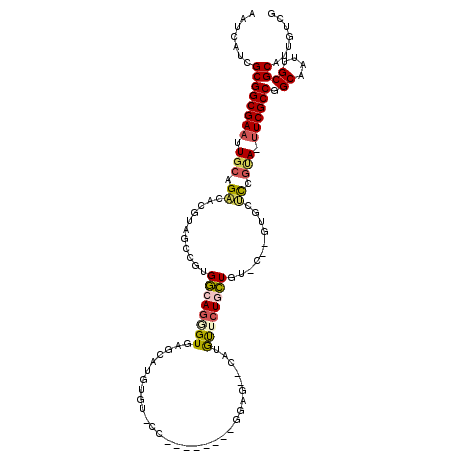
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,601,699 – 6,601,797 |
| Length | 98 |
| Max. P | 0.948792 |

| Location | 6,601,699 – 6,601,797 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6601699 98 + 22224390 AAUCAUCGCGGCGAAUUGCAGACACGUAGCCUUGGCAGAGUGAGCAUUU---------------------UGCUCUGCUAUUCGUGUCUAUCGUA-UUCGCCGGCAAUUGCGCAUUGUGA .....(((((((((((.(((((((((......((((((((..((...))---------------------..))))))))..)))))))...)))-)))))).((....)).....)))) ( -39.30) >DroSec_CAF1 16778 103 + 1 AAUCAUAGCGGCGAAUUGCAGACACGUAGCUGAGGCAGGGUGAGUAAGUAUUCC--------GGAG--CAUAUUCUGCU------GUGCUCCGUA-UUCGCCGGCAAUUGCGCAUUGAUG .((((..(((.(.((((((.........((....))..((((((((.......(--------((((--((((......)------))))))))))-)))))).))))))))))..)))). ( -37.41) >DroSim_CAF1 14151 108 + 1 AAUCAUAGCGGCGAAUUGCAGACACGUAGCUGAGGCAGGGUGAGGGUGAUUCCC--------GGAG--CAUAUUCUGCUCU-UGUGUGCUCCGUA-UUCGCCGGCAAUUGCGCAUUGUUG .....((((.(((((((((...(((.(.((....)).).)))..(((((....(--------((((--(((((........-.))))))))))..-.))))).)))))).)))...)))) ( -40.10) >DroEre_CAF1 17003 116 + 1 AUUCAUCGCGGCGAUUUGCAGA--CGGAGCCGUGUCAGUGUGCAUGUAUGUACGCGUAUUUGGUGGCACUUGCUAUUGUGUGC--GUGCUCCUAAUUUCGCCGGCAAAUGCGCAUUGUCG .......(((((..((((....--)))))))))(.((((((((((...(((..(((...((((..((((.(((......))).--))))..))))...)))..))).)))))))))).). ( -41.50) >DroYak_CAF1 16471 113 + 1 AUUCGUCGCGGCGAAUUGCAGU--UGUAGCCGUGUUAGUGUGAGCACGUGUGCCUGUAU---GUGGGACUUGUUCUUGUGUGC--GUGCACCGCAUUUCGCCGGCUAUUGCGCAUUGUCG .......((((((((.(((...--......((((((......)))))).((((.(((((---(..(((.....)))..)))))--).)))).))).)))))).((....))))....... ( -41.30) >consensus AAUCAUCGCGGCGAAUUGCAGACACGUAGCCGUGGCAGGGUGAGCAUGUGU_CC________GGAG__CAUGUUCUGCUGU_C__GUGCUCCGUA_UUCGCCGGCAAUUGCGCAUUGUCG .......((((((((.(((.((...........((((((((..............................))))))))..........)).))).)))))).((....))))....... (-17.59 = -17.83 + 0.24)

| Location | 6,601,699 – 6,601,797 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -11.64 |
| Energy contribution | -13.80 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6601699 98 - 22224390 UCACAAUGCGCAAUUGCCGGCGAA-UACGAUAGACACGAAUAGCAGAGCA---------------------AAAUGCUCACUCUGCCAAGGCUACGUGUCUGCAAUUCGCCGCGAUGAUU ...((.(((((....)).((((((-(..(.((((((((..((((.((((.---------------------....))))...((....))))))))))))))).)))))))))).))... ( -36.40) >DroSec_CAF1 16778 103 - 1 CAUCAAUGCGCAAUUGCCGGCGAA-UACGGAGCAC------AGCAGAAUAUG--CUCC--------GGAAUACUUACUCACCCUGCCUCAGCUACGUGUCUGCAAUUCGCCGCUAUGAUU .((((..((((....)).((((((-(.(((((((.------(......).))--))))--------)...........(((...((....))...)))......)))))))))..)))). ( -30.20) >DroSim_CAF1 14151 108 - 1 CAACAAUGCGCAAUUGCCGGCGAA-UACGGAGCACACA-AGAGCAGAAUAUG--CUCC--------GGGAAUCACCCUCACCCUGCCUCAGCUACGUGUCUGCAAUUCGCCGCUAUGAUU ...((..((((....)).((((((-(.((((.(((...-.(((((.....))--))).--------(((.....)))..................)))))))..)))))))))..))... ( -31.70) >DroEre_CAF1 17003 116 - 1 CGACAAUGCGCAUUUGCCGGCGAAAUUAGGAGCAC--GCACACAAUAGCAAGUGCCACCAAAUACGCGUACAUACAUGCACACUGACACGGCUCCG--UCUGCAAAUCGCCGCGAUGAAU ...((.(((((....)).(((((.....(((((..--((((..........))))..........((((......))))...........)))))(--....)...)))))))).))... ( -30.10) >DroYak_CAF1 16471 113 - 1 CGACAAUGCGCAAUAGCCGGCGAAAUGCGGUGCAC--GCACACAAGAACAAGUCCCAC---AUACAGGCACACGUGCUCACACUAACACGGCUACA--ACUGCAAUUCGCCGCGACGAAU ......(((((....)).((((((.((((((....--((............((.((..---.....)).)).((((..........))))))....--)))))).)))))))))...... ( -30.70) >consensus CAACAAUGCGCAAUUGCCGGCGAA_UACGGAGCAC__G_ACAGCAGAACAAG__CCAC________GG_ACACAUACUCACACUGCCACGGCUACGUGUCUGCAAUUCGCCGCGAUGAUU ...((.(((((....)).((((((...((((...........((((....................................))))............))))...))))))))).))... (-11.64 = -13.80 + 2.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:02 2006