
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,594,856 – 6,594,989 |
| Length | 133 |
| Max. P | 0.917108 |

| Location | 6,594,856 – 6,594,968 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -15.55 |
| Energy contribution | -17.92 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
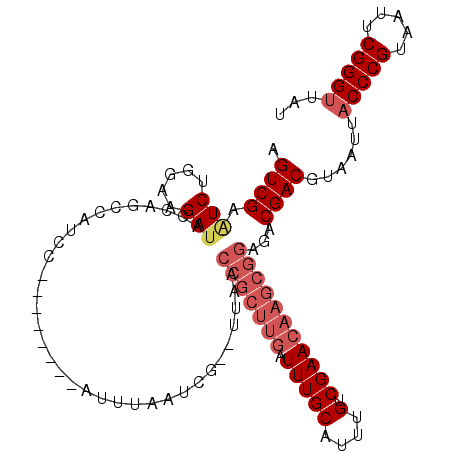
>X_DroMel_CAF1 6594856 112 + 22224390 AGUCGAAUCUGGAAGAUAUCAGCCAUCCAUACAUCCAUUUAAUCGCUUUAACCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAU .((((....((((.(........).))))......................(((((((.(((((....).)))))))))))...))))......(((((.....)))))... ( -29.20) >DroSim_CAF1 7218 112 + 1 AGUCGAGUCUGGAAGAUGCCAGGCAUCCAUCCAUCCAUUUAAUCGCUUUAACCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUANNNNNNNNNNNNNNNNN .((((.(..((((.(((((...)))))..))))..)...............(((((((.(((((....).)))))))))))...))))........................ ( -29.00) >DroEre_CAF1 10517 102 + 1 CGUCGACUCCUAAAGAUGCCAGCCAUCC--------AUUUUAUCG--UCAUCCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAU ((((..........((((.....)))).--------........(--((.((((((((.(((((....).))))))))))))))))))).....(((((.....)))))... ( -32.90) >DroYak_CAF1 9749 87 + 1 AGUCGAAUCUGGAAGAUAGCAGCCAACC--------AUUU-----------------AAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAU .((((..((((.......((((......--------....-----------------...)))).((((.....))))))))..))))......(((((.....)))))... ( -19.32) >consensus AGUCGAAUCUGGAAGAUACCAGCCAUCC________AUUUAAUCG__UUAACCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAU .((((.(((.....)))..................................(((((((.(((((....).)))))))))))...))))......(((((.....)))))... (-15.55 = -17.92 + 2.37)

| Location | 6,594,888 – 6,594,989 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
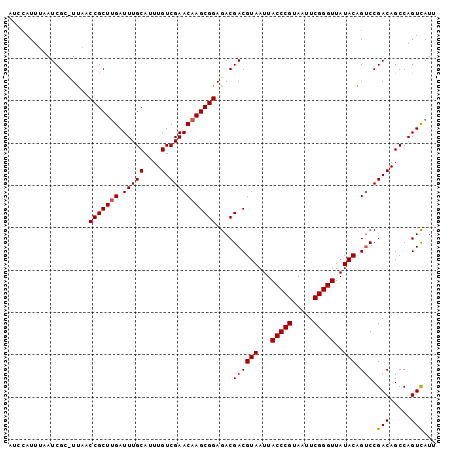
>X_DroMel_CAF1 6594888 101 + 22224390 AUCCAUUUAAUCGCUUUAACCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAUACAGACCGACAGCCAGUCAUU ..........(((((....(((((((.(((((....).))))))))))))).))).(((...(((((.....)))))..))).....(((.....)))... ( -26.80) >DroSec_CAF1 10444 101 + 1 AUCCAUUAAAUCGCCUUAACCGCUGGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAUACAGUCCGACAGCCAGUCAUU .....................(((((........((((((.((..(((....)).)(((...(((((.....)))))..))).)).))))))))))).... ( -26.10) >DroEre_CAF1 10545 95 + 1 ----AUUUUAUCG--UCAUCCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAUACGGUCCGACAGCCAGUUAUU ----......(((--((.((((((((.(((((....).))))))))))))))))).(((...(((((.....)))))..)))(((......)))....... ( -34.60) >consensus AUCCAUUUAAUCGC_UUAACCGCUUGAUUUGCAUUUGUCGAACAAGCGGAGACGACGUAAUUACCCGUAAUUCGGGUUAUACAGUCCGACAGCCAGUCAUU ...................(((((((.(((((....).)))))))))))....((((((...(((((.....)))))..))).))).(((.....)))... (-24.02 = -24.47 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:53 2006