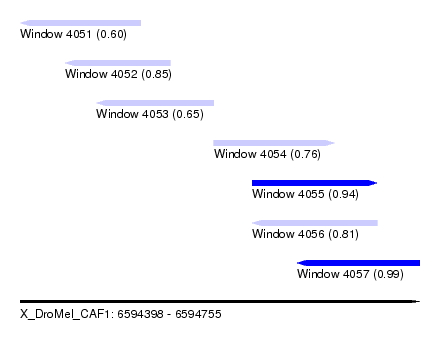
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,594,398 – 6,594,755 |
| Length | 357 |
| Max. P | 0.990674 |

| Location | 6,594,398 – 6,594,506 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.29 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.68 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594398 108 - 22224390 AGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUG-------GGAGAUUGCCACUGGUGCUGUUAGUUGCAUUAUUGAAUUGAUUGGAAGAAG ......(((((((.....((((((....)))))).....)))))))(((((((..((-------(.((...(((...))).)).))).))))))).................... ( -27.00) >DroSec_CAF1 9991 103 - 1 AGGAACUGGGAAUGUGUGGAACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUGCGAGAUGCGAGAUUGCCAC------------UUGCAUUAUUGAAUUGAUUGGAAGAAG ......(((((((.....((((((....)))))).....)))))))((((((((..((((.........))))..)------------))))))).................... ( -28.90) >DroEre_CAF1 10126 90 - 1 G----CAGAGGA--UGUGGGACUGGAGACGGUUCAUUUGAUUUCCGAAUA------G-------GGAGAUUGCUACGAG------UAGGUGCAUUAUCGAAAUGAUUGGACGGAG .----..((.((--(((.((((((....))))))....(((((((.....------.-------))))))).(((....------)))..))))).))................. ( -22.40) >consensus AGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUG_______GGAGAUUGCCAC__G______UAGUUGCAUUAUUGAAUUGAUUGGAAGAAG ...............(((((((((....))))))....(((((((...................)))))))..)))............((.(((((......))).)).)).... (-15.23 = -14.68 + -0.55)


| Location | 6,594,438 – 6,594,532 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594438 94 - 22224390 GAGCUAUUAUAGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUG-------GGAGAUUGCCA ............((.(((.(.((((..(.(..(((((((.....((((((....)))))).....)))))))..).)....))-------))).))).)). ( -25.80) >DroSec_CAF1 10019 101 - 1 GAGCUAUUAUGGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGAACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUGCGAGAUGCGAGAUUGCCA .........((((........)))).......(((((((.....((((((....)))))).....)))))))...((((..(((.....)))...)))).. ( -27.00) >DroSim_CAF1 6825 83 - 1 GAGCUAUUAUAGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUG------------------ ..((.......(((.......)))........(((((((.....((((((....)))))).....)))))))...))......------------------ ( -22.60) >DroEre_CAF1 10160 82 - 1 GAGCUAUUAUAGGGAAAUGUGCCCAAG----CAGAGGA--UGUGGGACUGGAGACGGUUCAUUUGAUUUCCGAAUA------G-------GGAGAUUGCUA ...........(((.......))).((----(......--....((((((....))))))....(((((((.....------.-------)))))))))). ( -23.60) >consensus GAGCUAUUAUAGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGAUUUCCAAAUGCGAGAUG_______GGAGAUUGCCA ............((((((...((((.......))))........((((((....)))))).....)))))).............................. (-18.46 = -18.77 + 0.31)

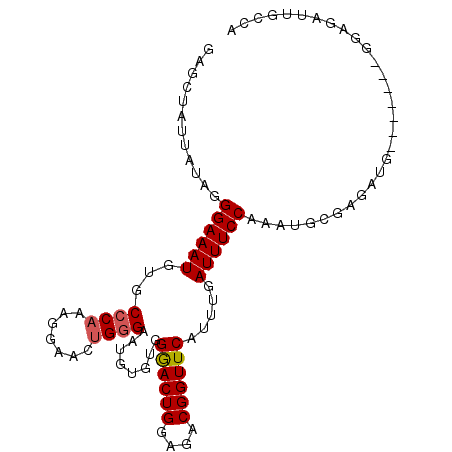
| Location | 6,594,466 – 6,594,571 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -22.29 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594466 105 - 22224390 UAUAUAUGCACAUCUGACAUUCCGCGA-UCGAAGCCGAUUGAGCUAUUAUAGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGA .......((((((((.(..((((.(((-(((....))))))..........(((.......)))...)))).).)).)))))).((((((....))))))...... ( -31.40) >DroSec_CAF1 10054 87 - 1 ------------------AUUCCGCGA-UCGAAGCCGAUUGAGCUAUUAUGGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGAACUGGAGACGGUUCAUUUGA ------------------(((((.(((-(((....))))))........((((........)))).........))))).....((((((....))))))...... ( -26.40) >DroSim_CAF1 6842 103 - 1 U--AUAUGCACAUCUGACAUUCCGCGA-UCGAAGCCGAUUGAGCUAUUAUAGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGA .--....((((((((.(..((((.(((-(((....))))))..........(((.......)))...)))).).)).)))))).((((((....))))))...... ( -31.40) >DroEre_CAF1 10182 91 - 1 ---------AGAUCUGACAUUCCGCGAUUCGAAGCUGAUUGAGCUAUUAUAGGGAAAUGUGCCCAAG----CAGAGGA--UGUGGGACUGGAGACGGUUCAUUUGA ---------.......(((((((((.......((((.....))))......(((.......)))..)----).).)))--))).((((((....))))))...... ( -24.70) >consensus _________ACAUCUGACAUUCCGCGA_UCGAAGCCGAUUGAGCUAUUAUAGGGAAAUGUGCCCAAAGGAACUGGGAAUGUGUGGGACUGGAGACGGUUCAUUUGA ................(((((((((...(((....)))....)).......(((.......)))..........)))))))...((((((....))))))...... (-22.29 = -22.98 + 0.69)

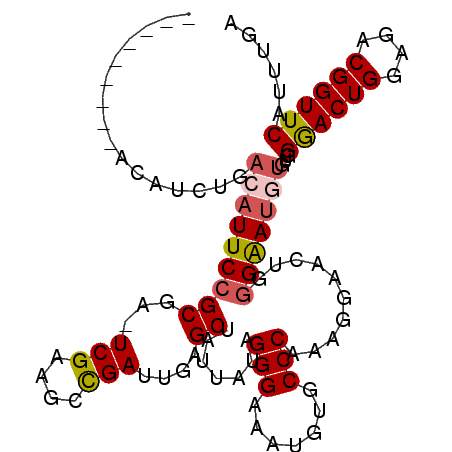
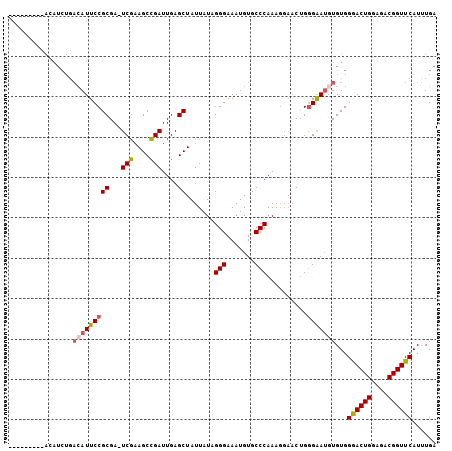
| Location | 6,594,571 – 6,594,679 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -16.24 |
| Energy contribution | -15.48 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594571 108 + 22224390 UGUAUGUAUUGUAU------AUGCGAGCAAUUAGAAAGGUUGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG------CUGGCAUCCAGCUUAAUUGUUUGACCACUCU .(((((((...)))------))))((((((((.....)))))))).((((...(((((((((((........((......))(------((((...))))))))))))))))..)))).. ( -31.40) >DroSec_CAF1 10141 97 + 1 ----------AUAU------AUGCGAGCAAUUAGAAAGGUUGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG------CCGGCAUCCAGCUUAAUUGU-UGACCACUCU ----------....------....((((((((.....)))))))).(((((((((((..((((((....))))))...))).)------.)))))))((((......))-))........ ( -24.10) >DroSim_CAF1 6945 107 + 1 UGUACGAGCAAUAU------AUGCGAGCAAUUAGAAAGGUUGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG------CUGGCAUCCAGCUUAAUUGU-UGACCACUCU .....((((((((.------..((((((((((.....)))))))).(((((((((((..((((((....))))))...))).)------).))))))..)).....)))-))....))). ( -26.80) >DroEre_CAF1 10273 92 + 1 ----------AUAUGAGUGUUUGCGAGUAAUUAGAAAGGUUGCUCAGGUGUCGCAGAUGGUUGACUUUAAUUGAUAACUCCCG------CUGGCAUCCAGCUUAAUUG------------ ----------....((((((((((((((((((.....)))))))).(....))))))).((..(......)..)).))))..(------((((...))))).......------------ ( -21.50) >DroYak_CAF1 9453 104 + 1 ----------AUAU------AUGCGAGUAAUUAGAAAGGUUGCUCAGGUGUCGCAGAUGAUUAAUUUUAAUUGAUAACUCUCGCAUAAGCUGGCAGCCAGCUUAAUUGCUCGACUACUCU ----------....------...(((((((((((...((((((.(((.(...(((((..((((((....))))))...))).))...).)))))))))...)))))))))))........ ( -32.50) >consensus __________AUAU______AUGCGAGCAAUUAGAAAGGUUGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG______CUGGCAUCCAGCUUAAUUGU_UGACCACUCU ......................((((((((((.....)))))))).(((((((.....(((((....)))))((......))........))))).)).))................... (-16.24 = -15.48 + -0.76)

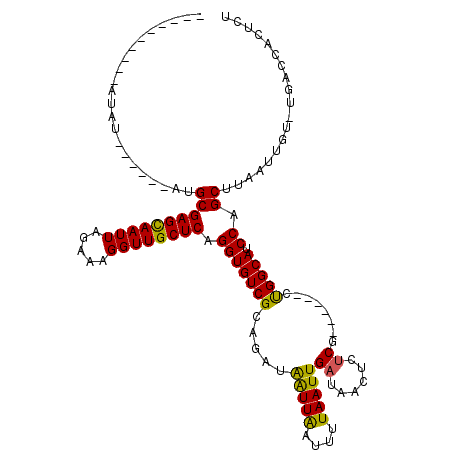

| Location | 6,594,605 – 6,594,717 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -20.61 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594605 112 + 22224390 UGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG------CUGGCAUCCAGCUUAAUUGUUUGACCACUCUAAUUAUGCCAGGCCAUCUGGCAUCUUGAGCGAU--AUUUA .(((((((((...(((((((((((........((......))(------((((...))))))))))))))))..))))......(((((((.....)))))))..)))))...--..... ( -35.00) >DroSec_CAF1 10165 111 + 1 UGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG------CCGGCAUCCAGCUUAAUUGU-UGACCACUCUAAUUAUGCCAGGCCAUCUGGCAUCUUGAGCGAU--AUUUA .((((((((((((((((..((((((....))))))...))).)------.)))))))((((......))-))............(((((((.....)))))))..)))))...--..... ( -31.10) >DroSim_CAF1 6979 111 + 1 UGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG------CUGGCAUCCAGCUUAAUUGU-UGACCACUCUAAUUAUGCCAGGCCAUCUGGCAUCUUGAGCGAU--AUUUA .(((((((.(((...(((((((((........((......))(------((((...)))))))))))))-))))..........(((((((.....))))))))))))))...--..... ( -31.80) >DroEre_CAF1 10303 95 + 1 UGCUCAGGUGUCGCAGAUGGUUGACUUUAAUUGAUAACUCCCG------CUGGCAUCCAGCUUAAUUG-----------------AGCCAGACCAUGUGGCCACUUGAGUGAU--AUUUG .(((((((((.(.((.((((((...(((((((((........(------((((...))))))))))))-----------------))...)))))).))).)))))))))...--..... ( -33.90) >DroYak_CAF1 9477 120 + 1 UGCUCAGGUGUCGCAGAUGAUUAAUUUUAAUUGAUAACUCUCGCAUAAGCUGGCAGCCAGCUUAAUUGCUCGACUACUCUAAUUAUGCCAGGCCAUCUGGCAUCUUGAGUGAUAUAUUUA .(((((((.((((((((..((((((....))))))...))).)).((((((((...)))))))).......)))..........(((((((.....)))))))))))))).......... ( -36.80) >consensus UGCUCAGGUGUCGCAGAUAAUUAAUUUUAAUUGAUAACUCUCG______CUGGCAUCCAGCUUAAUUGU_UGACCACUCUAAUUAUGCCAGGCCAUCUGGCAUCUUGAGCGAU__AUUUA .((((((((((..(((((.((((((....))))))..............((((((..............................))))))...)))))))).))))))).......... (-20.61 = -21.05 + 0.44)

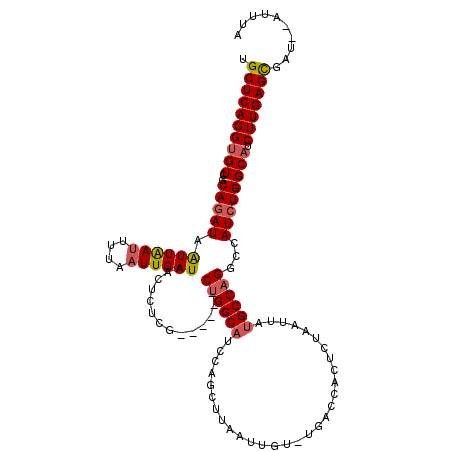

| Location | 6,594,605 – 6,594,717 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -20.14 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594605 112 - 22224390 UAAAU--AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCAAACAAUUAAGCUGGAUGCCAG------CGAGAGUUAUCAAUUAAAAUUAAUUAUCUGCGACACCUGAGCA .....--...(((((.((((((((.....)))))))..........(((..........(((((...))))------)((.(((((...........))))).))...))).).))))). ( -30.20) >DroSec_CAF1 10165 111 - 1 UAAAU--AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCA-ACAAUUAAGCUGGAUGCCGG------CGAGAGUUAUCAAUUAAAAUUAAUUAUCUGCGACACCUGAGCA .....--...(((((.(.((((((((((((.(((((...(((..((((..-...))))..))).)))))))------)((......)).............)))))).).))).))))). ( -30.20) >DroSim_CAF1 6979 111 - 1 UAAAU--AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCA-ACAAUUAAGCUGGAUGCCAG------CGAGAGUUAUCAAUUAAAAUUAAUUAUCUGCGACACCUGAGCA .....--...(((((.((((((((.....))))))).......((.((..-(.(((((((((((...))))------)((......))........)))))).)..)).)).).))))). ( -30.70) >DroEre_CAF1 10303 95 - 1 CAAAU--AUCACUCAAGUGGCCACAUGGUCUGGCU-----------------CAAUUAAGCUGGAUGCCAG------CGGGAGUUAUCAAUUAAAGUCAACCAUCUGCGACACCUGAGCA .....--....((((.(((.(((.(((((.(((((-----------------.......(((((...))))------)..((....))......)))))))))).)).).))).)))).. ( -26.20) >DroYak_CAF1 9477 120 - 1 UAAAUAUAUCACUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUAGUCGAGCAAUUAAGCUGGCUGCCAGCUUAUGCGAGAGUUAUCAAUUAAAAUUAAUCAUCUGCGACACCUGAGCA ........((((((...(((((((.....))))))).....)))).((((.(((..((((((((...)))))))))))((..((((...........))))..))..))))....))... ( -34.00) >consensus UAAAU__AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCA_ACAAUUAAGCUGGAUGCCAG______CGAGAGUUAUCAAUUAAAAUUAAUUAUCUGCGACACCUGAGCA ..........(((((.(.((((((((((.(((((((...(((..((((......))))..))).))))))).......((......))............))))))).).)).)))))). (-20.14 = -22.10 + 1.96)

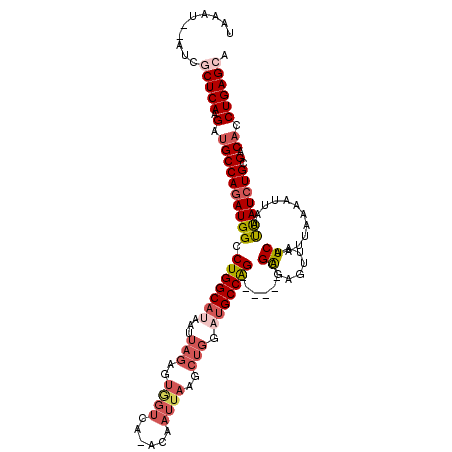
| Location | 6,594,645 – 6,594,755 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6594645 110 - 22224390 CACUAGCCCUAGUCG--CAGAUUGGGCAAUAAAUUACUCAUAAAU--AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCAAACAAUUAAGCUGGAUGCCAG------CGA .....((((.((((.--..))))))))..................--(((((((...(((((((.....))))))).....)))))))...........(((((...))))------).. ( -34.20) >DroSec_CAF1 10205 109 - 1 CACUAGCCCUAGUCG--CAGAUUGGGCAAUAAAUUACUCAUAAAU--AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCA-ACAAUUAAGCUGGAUGCCGG------CGA .....((((.((((.--..))))))))..................--(((((((...(((((((.....))))))).....)))))))..-........(((((...))))------).. ( -33.50) >DroSim_CAF1 7019 109 - 1 CACUAGCCCUAGUCG--CAGAUUGGGCAAUAAAUUACUCAUAAAU--AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCA-ACAAUUAAGCUGGAUGCCAG------CGA .....((((.((((.--..))))))))..................--(((((((...(((((((.....))))))).....)))))))..-........(((((...))))------).. ( -34.20) >DroYak_CAF1 9517 120 - 1 CAGCGGCCCUAGUCGCCCAGAUUGGGCAAUAAAUUGCACAUAAAUAUAUCACUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUAGUCGAGCAAUUAAGCUGGCUGCCAGCUUAUGCGA ...((((.......((((.....)))).......................((((...(((((((.....))))))).....)))).)))).(((..((((((((...))))))))))).. ( -39.70) >consensus CACUAGCCCUAGUCG__CAGAUUGGGCAAUAAAUUACUCAUAAAU__AUCGCUCAAGAUGCCAGAUGGCCUGGCAUAAUUAGAGUGGUCA_ACAAUUAAGCUGGAUGCCAG______CGA ..((((((((((((.....))))))).....................(((((((...(((((((.....))))))).....)))))))...........)))))................ (-31.06 = -31.00 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:51 2006