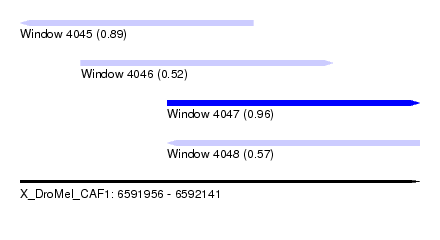
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,591,956 – 6,592,141 |
| Length | 185 |
| Max. P | 0.962334 |

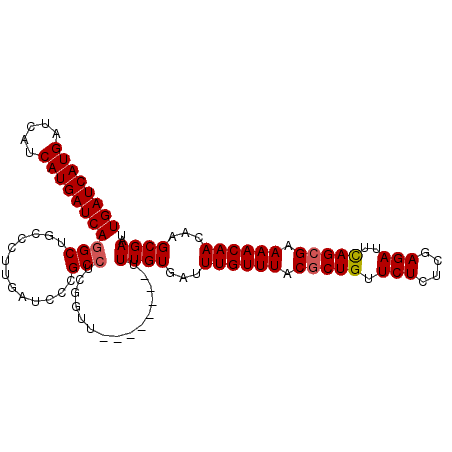
| Location | 6,591,956 – 6,592,064 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.51 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -24.41 |
| Energy contribution | -24.85 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6591956 108 - 22224390 UGAUCAUGAUCAUCAUGAUCACGCUACCCUUGAUCCCGCCUCGUUUUUUUUUUUUUUGUGAUUUGUUUACGCUGUUCUCUCGAGAUUUAGCGAAAACAACAAGCGAUU ((((((((.....))))))))((((...........(((..................)))..((((((.(((((.(((....)))..))))).))))))..))))... ( -26.57) >DroSec_CAF1 7636 100 - 1 UGAUCAUGAUCAUCAUGAUCAGGCUGCCCUUGAUCCCGCCUCGGUU--------UUUGUGAUUUGUUUACCCUGUUCUCUCGAGAUUCAGCGAAAACAACAAGCGAAU ((((((((.....))))))))(((.............)))......--------(((((...((((((.(.(((.(((....)))..))).).))))))...))))). ( -24.62) >DroSim_CAF1 4444 100 - 1 UGAUCAUGAUCAUCAUGAUCAGGCUGCCCUUGAUCCCGCCUCGGUU--------UUUGUGAUUUGUUUACGCUGUUCUCUCGAGAGUCAGCGAAAACAACAAGCGAAU ((((((((.....))))))))(((.............)))......--------(((((...((((((.(((((((((....)))).))))).))))))...))))). ( -32.22) >consensus UGAUCAUGAUCAUCAUGAUCAGGCUGCCCUUGAUCCCGCCUCGGUU________UUUGUGAUUUGUUUACGCUGUUCUCUCGAGAUUCAGCGAAAACAACAAGCGAAU ((((((((.....))))))))(((.............)))...............((((...((((((.(((((.(((....)))..))))).))))))...)))).. (-24.41 = -24.85 + 0.45)


| Location | 6,591,984 – 6,592,101 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -18.46 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6591984 117 + 22224390 AGAGAACAGCGUAAACAAAUCACAAAAAAAAAAAAAACGAGGCGGGAUCAAGGGUAGCGUGAUCAUGAUGAUCAUGAUCAUCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG--- ........................................(((..(((((.((((.(((((....(((((((....)))))))..))))).))))....))))).))).........--- ( -29.70) >DroSec_CAF1 7664 109 + 1 AGAGAACAGGGUAAACAAAUCACAAA--------AACCGAGGCGGGAUCAAGGGCAGCCUGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCCGCCGAAUGGCCG--- .........(((.......((.....--------....))(((((.((((((.((((((((((((((.....)))))))...)))).)))..)).....))))))))).....))).--- ( -33.00) >DroSim_CAF1 4472 109 + 1 AGAGAACAGCGUAAACAAAUCACAAA--------AACCGAGGCGGGAUCAAGGGCAGCCUGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG--- ..........................--------..(((.(((..(((((((.((((((((((((((.....)))))))...)))).)))..)).....))))).)))...)))...--- ( -30.50) >DroEre_CAF1 7828 97 + 1 AGAGAACAGCGCAAGCGAAUCAUA--------------GAGGCGGGAUCAAGGGCAGCA---------UGGUCAUGAUCAGCGAGCAUGCAGCUCAAGAUGCUUAGCCGAGUGGCCGAGG ........((....))...((((.--------------..(((((.(((..((((.(((---------((.((.........)).))))).))))..))).))..)))..))))...... ( -30.10) >DroYak_CAF1 6987 94 + 1 AGAGGAGAACGUAAACAAAUCAUA--------------GAGGCGGGAUCAAGGGCAGCA---------UGAUCAUGAUCAGCACGCAUGCAGCUCGAGAUGAUCAGCUGAGUGGCCG--- ...((.....(....)...((((.--------------..(((..(((((.((((.(((---------((....((.....))..))))).))))....))))).)))..)))))).--- ( -22.90) >consensus AGAGAACAGCGUAAACAAAUCACAAA________AA_CGAGGCGGGAUCAAGGGCAGCAUGAUCAUGAUGAUCAUGAUCACCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG___ ........................................(((..(((((.((((.(((((.......((((....)))).....))))).))))....))))).)))............ (-18.46 = -19.14 + 0.68)



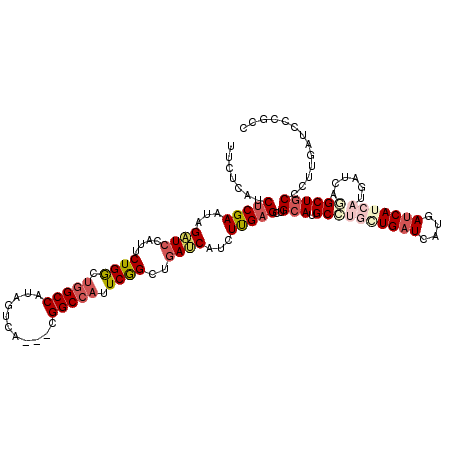
| Location | 6,592,024 – 6,592,141 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.21 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6592024 117 + 22224390 GGCGGGAUCAAGGGUAGCGUGAUCAUGAUGAUCAUGAUCAUCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAA (((..(((((.((((.(((((....(((((((....)))))))..))))).))))....))))).)))(..((((((---(....)))))))..).(((((....))))).......... ( -43.20) >DroSec_CAF1 7696 117 + 1 GGCGGGAUCAAGGGCAGCCUGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCCGCCGAAUGGCCG---UGACUAUGGCCAACCAGAAUGGAUCUAUUCGAGAUGAGAA (((((.((((((.((((((((((((((.....)))))))...)))).)))..)).....)))))))))...((((((---(....)))))))....(((((....))))).......... ( -44.10) >DroSim_CAF1 4504 117 + 1 GGCGGGAUCAAGGGCAGCCUGAUCAUGAUGAUCAUGAUCACCAGGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG---UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAA (((..(((((((.((((((((((((((.....)))))))...)))).)))..)).....))))).)))(..((((((---(....)))))))..).(((((....))))).......... ( -42.60) >DroEre_CAF1 7854 111 + 1 GGCGGGAUCAAGGGCAGCA---------UGGUCAUGAUCAGCGAGCAUGCAGCUCAAGAUGCUUAGCCGAGUGGCCGAGGUGACUGCGGCUGGCCAGAACUGAGCUAUUCGCGAAAGGAA (((((.(((..((((.(((---------((.((.........)).))))).))))..))).))..)))(((((((((((....)).)))))(((((....)).))))))).(....)... ( -42.00) >DroYak_CAF1 7013 108 + 1 GGCGGGAUCAAGGGCAGCA---------UGAUCAUGAUCAGCACGCAUGCAGCUCGAGAUGAUCAGCUGAGUGGCCG---UGACUGUGGCCAGUCAGAAUGGAACUAUUCGAGCAAAGGA .(((.(((((.(..(....---------.)..).)))))....))).....(((((((..(.((..((((.((((((---......)))))).))))....)).)..)))))))...... ( -39.40) >consensus GGCGGGAUCAAGGGCAGCAUGAUCAUGAUGAUCAUGAUCACCAAGCAUGCAGCUCAAGAUGAUCAGCCGAAUGGCCG___UGACUAUGGCCAGCCAGAAUGGAUCUAUUCGAGAUGAGAA (((..(((((.((((.(((((.......((((....)))).....))))).))))....))))).)))(..((((((.........))))))..).(((((....))))).......... (-28.04 = -28.64 + 0.60)



| Location | 6,592,024 – 6,592,141 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.21 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -21.22 |
| Energy contribution | -23.26 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6592024 117 - 22224390 UUCUCAUCUCGAAUAGAUCCAUUCUGGCUGGCCAUAGUCA---CGGCCAUUCGGCUGAUCAUCUUGAGCUGCAUGCUUGAUGAUCAUGAUCAUCAUGAUCACGCUACCCUUGAUCCCGCC ..........(((((((.....))))..(((((.......---.))))))))(((.(((((..(..(((.....)))..)((((((((.....)))))))).........)))))..))) ( -34.50) >DroSec_CAF1 7696 117 - 1 UUCUCAUCUCGAAUAGAUCCAUUCUGGUUGGCCAUAGUCA---CGGCCAUUCGGCGGAUCAUCUUGAGCUGCAUGCCUGGUGAUCAUGAUCAUCAUGAUCAGGCUGCCCUUGAUCCCGCC ...(((.(((((...(((((...(((..(((((.......---.)))))..))).)))))...)))))..(((.((((...(((((((.....))))))))))))))...)))....... ( -40.70) >DroSim_CAF1 4504 117 - 1 UUCUCAUCUCGAAUAGAUCCAUUCUGGCUGGCCAUAGUCA---CGGCCAUUCGGCUGAUCAUCUUGAGCUGCAUGCCUGGUGAUCAUGAUCAUCAUGAUCAGGCUGCCCUUGAUCCCGCC ...(((.(((((...((((....(((..(((((.......---.)))))..)))..))))...)))))..(((.((((...(((((((.....))))))))))))))...)))....... ( -37.10) >DroEre_CAF1 7854 111 - 1 UUCCUUUCGCGAAUAGCUCAGUUCUGGCCAGCCGCAGUCACCUCGGCCACUCGGCUAAGCAUCUUGAGCUGCAUGCUCGCUGAUCAUGACCA---------UGCUGCCCUUGAUCCCGCC ........(((...(((((((.....((.(((((.(((..(....)..))))))))..))...)))))))(((((.(((.......))).))---------)))............))). ( -30.50) >DroYak_CAF1 7013 108 - 1 UCCUUUGCUCGAAUAGUUCCAUUCUGACUGGCCACAGUCA---CGGCCACUCAGCUGAUCAUCUCGAGCUGCAUGCGUGCUGAUCAUGAUCA---------UGCUGCCCUUGAUCCCGCC ......((((((.....((....((((.(((((.......---.))))).))))..)).....)))))).(((((((((.....))))..))---------)))................ ( -29.90) >consensus UUCUCAUCUCGAAUAGAUCCAUUCUGGCUGGCCAUAGUCA___CGGCCAUUCGGCUGAUCAUCUUGAGCUGCAUGCCUGCUGAUCAUGAUCAUCAUGAUCAGGCUGCCCUUGAUCCCGCC .......(((((...((((....((((.(((((...........))))).))))..))))...)))))..(((.((((((((((....)))))))......))))))............. (-21.22 = -23.26 + 2.04)

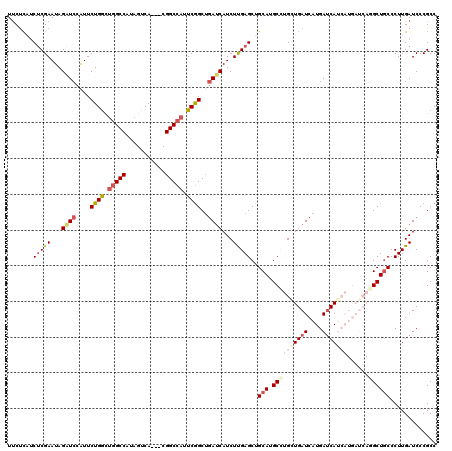
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:44 2006