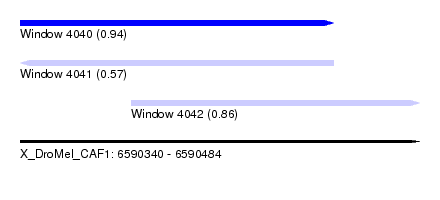
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,590,340 – 6,590,484 |
| Length | 144 |
| Max. P | 0.941823 |

| Location | 6,590,340 – 6,590,453 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.45 |
| Mean single sequence MFE | -35.59 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.84 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6590340 113 + 22224390 AAUCGGCUGGCGAUCAAAAAUAAGACUGCGGAUCAAACCGGUCGAAGGAGGCGGUGCUCCACCGCUCACCACUCACCGAUGGCCACUGACCAGUGACCACUCACCACUAA-------CCA ..((((.((((.(((........(((((.(.......))))))...((.(((((((...)))))))..)).......))).))))))))...((((....))))......-------... ( -35.70) >DroSec_CAF1 6022 110 + 1 AAUCGGCCGGCGAUCAAAAAUAAGACUGCGGAUCAAACCGGUCGAAGGAGGCGGUACUCCACCGCUCACCACUCACCGAUGGCCACUGACCAGUGGCCAGUGACCACUCA---------- ..((((((((.((((...............))))...))))))))..((((((((.....))))).......((((...((((((((....))))))))))))...))).---------- ( -43.86) >DroEre_CAF1 6145 119 + 1 AAUCGGCCGGCGAUCAACAAUAAGACUGCGGAUCAAACCGGUCGA-GAAGGCGGUGCUCCACCGCUCACCACUGACCACUGACCACUGACCAGUCACCAGCCACCACUCACCACUGACCA ..((((((((.((((..((.......))..))))...))))))))-((.(((((((...))))))).....(((...((((.........))))...))).......))........... ( -32.70) >DroYak_CAF1 5216 99 + 1 AAUCGGCCGGCGAUCAAAAAUAAGACUGCGGAUCAAACCGGUCGG-GAAGGCGUUGCUCCACCGCUCACCACUGACCACUGGCCACUGGCCA----CCAACCA---C------------- ....((((((.(..((.............((......))((((((-...((.(..((......)).).)).))))))..))..).)))))).----.......---.------------- ( -30.10) >consensus AAUCGGCCGGCGAUCAAAAAUAAGACUGCGGAUCAAACCGGUCGA_GAAGGCGGUGCUCCACCGCUCACCACUCACCAAUGGCCACUGACCAGUGACCAGCCACCACUCA__________ ..((((((((.((((...............))))...)))))))).(..(((((((...)))))))..)..........(((.((((....)))).)))..................... (-25.46 = -26.84 + 1.38)


| Location | 6,590,340 – 6,590,453 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.45 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -32.05 |
| Energy contribution | -32.68 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6590340 113 - 22224390 UGG-------UUAGUGGUGAGUGGUCACUGGUCAGUGGCCAUCGGUGAGUGGUGAGCGGUGGAGCACCGCCUCCUUCGACCGGUUUGAUCCGCAGUCUUAUUUUUGAUCGCCAGCCGAUU .((-------(...(((((((((((((((....)))))))))(((((((.((.(.((((((...))))))).))))).)))).........................))))))))).... ( -43.40) >DroSec_CAF1 6022 110 - 1 ----------UGAGUGGUCACUGGCCACUGGUCAGUGGCCAUCGGUGAGUGGUGAGCGGUGGAGUACCGCCUCCUUCGACCGGUUUGAUCCGCAGUCUUAUUUUUGAUCGCCGGCCGAUU ----------....((((((((((((...))))))))))))(((((...((((((.((((((....)))))......((((((......)))..)))........).))))))))))).. ( -46.20) >DroEre_CAF1 6145 119 - 1 UGGUCAGUGGUGAGUGGUGGCUGGUGACUGGUCAGUGGUCAGUGGUCAGUGGUGAGCGGUGGAGCACCGCCUUC-UCGACCGGUUUGAUCCGCAGUCUUAUUGUUGAUCGCCGGCCGAUU ((..((((.(..(((....)))..).))))..)).(((((.((((((((..((((((.(((((..((((.....-.....))))....))))).).)))))..)))))))).)))))... ( -48.50) >DroYak_CAF1 5216 99 - 1 -------------G---UGGUUGG----UGGCCAGUGGCCAGUGGUCAGUGGUGAGCGGUGGAGCAACGCCUUC-CCGACCGGUUUGAUCCGCAGUCUUAUUUUUGAUCGCCGGCCGAUU -------------.---(((((((----(((((...))))...((((((..((((((.(((((.(((.(((...-......)))))).))))).).)))))..))))))))))))))... ( -40.40) >consensus __________UGAGUGGUGACUGGUCACUGGUCAGUGGCCAGCGGUCAGUGGUGAGCGGUGGAGCACCGCCUCC_UCGACCGGUUUGAUCCGCAGUCUUAUUUUUGAUCGCCGGCCGAUU ...................((((((.....))))))((((.((((((((..((((((.(((((.....(((..........)))....))))).).)))))..)))))))).)))).... (-32.05 = -32.68 + 0.63)


| Location | 6,590,380 – 6,590,484 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.82 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -16.14 |
| Energy contribution | -18.70 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6590380 104 + 22224390 GUCGAAGGAGGCGGUGCUCCACCGCUCACCACUCACCGAUGGCCACUGACCAGUGACCACUCACCACUAA-------CCAU--AC-------CACUCAGCAAAUCCUGCGGCAAAUCCGA ((((.(((((((((((...)))))))...........(((((.((((....)))).))).))........-------....--..-------...........)))).))))........ ( -29.20) >DroSec_CAF1 6062 99 + 1 GUCGAAGGAGGCGGUACUCCACCGCUCACCACUCACCGAUGGCCACUGACCAGUGGCCAGUGACCACUCA--------------C-------CACUUACCAAAUCCUGCGGCAAAUCCGA ((((.((((((((((.....))))))......((((...((((((((....)))))))))))).......--------------.-------...........)))).))))........ ( -36.00) >DroEre_CAF1 6185 119 + 1 GUCGA-GAAGGCGGUGCUCCACCGCUCACCACUGACCACUGACCACUGACCAGUCACCAGCCACCACUCACCACUGACCACUGACCACCCAUCAUCCACCAAAUCCUGCGACAAAUCCGA (((((-(..(((((((...))))))).............(((....((..((((...(((.............)))...))))..))....)))...........)).))))........ ( -20.32) >DroYak_CAF1 5256 84 + 1 GUCGG-GAAGGCGUUGCUCCACCGCUCACCACUGACCACUGGCCACUGGCCA----CCAACCA---C----------------------------UCACCAAAUCCUGCGGCAAAUCCCA ...((-((.((.......)).(((((((....)))....(((((...)))))----.......---.----------------------------............))))....)))). ( -19.10) >consensus GUCGA_GAAGGCGGUGCUCCACCGCUCACCACUCACCAAUGGCCACUGACCAGUGACCAGCCACCACUCA______________C_______CACUCACCAAAUCCUGCGGCAAAUCCGA ((((.(((((((((((...))))))).............(((.((((....)))).)))............................................)))).))))........ (-16.14 = -18.70 + 2.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:39 2006