
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,589,217 – 6,589,456 |
| Length | 239 |
| Max. P | 0.956919 |

| Location | 6,589,217 – 6,589,336 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.34 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -24.95 |
| Energy contribution | -26.32 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6589217 119 + 22224390 AAAAAUAUGUAAGAAAAAAAAUGAAUUGGU-UUGGUGUCAGCGCAUUUUCUGGCCGGCCAAUUCGACGUGUUGAUUUCCACCAGCCAGAAUUGCGCCGCAAACACCAUUAACACGACCGC ...........................(((-((((((((.(((((..(((((((.((.....((((....))))......)).))))))).))))).)...)))))).......)))).. ( -34.31) >DroSec_CAF1 4796 118 + 1 A-AAAUAUGUAAGAAAAAAAAUGAAUUGGU-UGGGUGUCAGCGCAUUUUCUGGCCGUCCAAUUCGACGUGUUGAUUUCCACCAGCCAGAAUUGCGCCGCAAACACCAUUAACACGACCGC .-.........................(((-((((((((.(((((..(((((((.(((......)))(((........)))..))))))).))))).)...))))).......))))).. ( -33.11) >DroEre_CAF1 4855 114 + 1 A-AAAUAUGUAAGAAAAAA-----AUUGGUGUUGAAGUAAACGCAUUUUCUGGCCGGCCAAUUCGGCGUGUUGAUUUCUACCAGCCAGAAUUGCGCCGCAAACACCAUUAGCACGACCGC .-.................-----..(((((((...((...((((..(((((((.((.....((((....))))......)).))))))).))))..)).)))))))............. ( -29.70) >DroYak_CAF1 3917 117 + 1 A-AAAUAUGUAAGAAAAAAA--GAAUUGGUUUCGGUGCAAACGCAUUUUCUGGCCGGCCAAUUGGACGUGUUGAUUUCUACCAGCCAGAAUUGCGCCGCCAACACCGUUAACACGACCGC .-..................--...((((((.(((((....((((..(((((((.((..(((..(.....)..)))....)).))))))).)))).......)))))..))).))).... ( -27.70) >consensus A_AAAUAUGUAAGAAAAAAA__GAAUUGGU_UUGGUGUAAACGCAUUUUCUGGCCGGCCAAUUCGACGUGUUGAUUUCCACCAGCCAGAAUUGCGCCGCAAACACCAUUAACACGACCGC .........................(((((..((((((..(((((..(((((((.((.....((((....))))......)).))))))).))))).....))))))...)).))).... (-24.95 = -26.32 + 1.37)


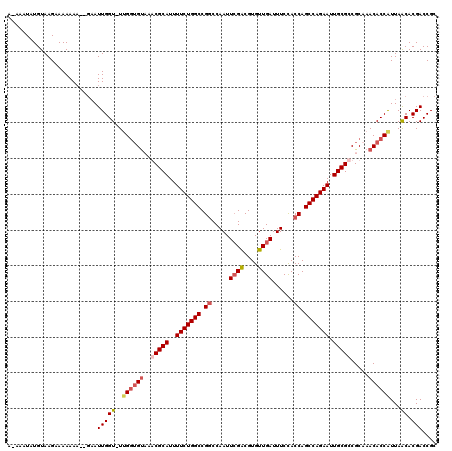
| Location | 6,589,217 – 6,589,336 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.34 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.13 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6589217 119 - 22224390 GCGGUCGUGUUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUGGAAAUCAACACGUCGAAUUGGCCGGCCAGAAAAUGCGCUGACACCAA-ACCAAUUCAUUUUUUUUCUUACAUAUUUUU ......(.(((..((((((...(((((((.(((((((((((.((..((........)).)).)))))))))))..))))))))))))))-)))........................... ( -39.50) >DroSec_CAF1 4796 118 - 1 GCGGUCGUGUUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUGGAAAUCAACACGUCGAAUUGGACGGCCAGAAAAUGCGCUGACACCCA-ACCAAUUCAUUUUUUUUCUUACAUAUUU-U ..(((.(.......(((((...(((((((.(((((((((((....))))...((((......)))))))))))..))))))))))))).-))).........................-. ( -37.31) >DroEre_CAF1 4855 114 - 1 GCGGUCGUGCUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUAGAAAUCAACACGCCGAAUUGGCCGGCCAGAAAAUGCGUUUACUUCAACACCAAU-----UUUUUUCUUACAUAUUU-U (((....)))...(((((((...((((((.(((((((((((..((.((........)).)).)))))))))))..))))))......)))))))..-----.................-. ( -34.50) >DroYak_CAF1 3917 117 - 1 GCGGUCGUGUUAACGGUGUUGGCGGCGCAAUUCUGGCUGGUAGAAAUCAACACGUCCAAUUGGCCGGCCAGAAAAUGCGUUUGCACCGAAACCAAUUC--UUUUUUUCUUACAUAUUU-U ..(((.((....))(((((....((((((.(((((((((((...(((...........))).)))))))))))..)))))).)))))...))).....--..................-. ( -35.10) >consensus GCGGUCGUGUUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUAGAAAUCAACACGUCGAAUUGGCCGGCCAGAAAAUGCGCUGACACCAA_ACCAAUUC__UUUUUUUCUUACAUAUUU_U ..(((........((((((...(((((((.(((((((((((.((...............)).)))))))))))..)))))))))))))..)))........................... (-31.76 = -32.13 + 0.37)


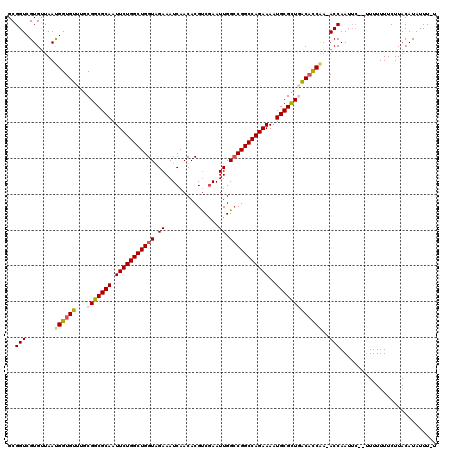
| Location | 6,589,256 – 6,589,376 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -35.08 |
| Energy contribution | -34.34 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6589256 120 - 22224390 CAAACUCAUUUAUUUGUAGCCAAGGCACUGUGGUACGUUUGCGGUCGUGUUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUGGAAAUCAACACGUCGAAUUGGCCGGCCAGAAAAUGCGC ..................(((.(((((((((..((((........))))...)))))))))..)))(((.(((((((((((.((..((........)).)).)))))))))))..))).. ( -40.40) >DroSec_CAF1 4834 120 - 1 UAAACUCAUUUACUUGUAGCUAAGGCACUGCGGUACGUUUGCGGUCGUGUUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUGGAAAUCAACACGUCGAAUUGGACGGCCAGAAAAUGCGC ((((((((((.((.....((....))(((((((.....)))))))...)).))))).)))))..(((((.(((((((((((....))))...((((......)))))))))))..))))) ( -39.70) >DroEre_CAF1 4889 120 - 1 AAAACUCAUUUACUUGUAGCCAGGGCAUUGCGGUACGUUUGCGGUCGUGCUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUAGAAAUCAACACGCCGAAUUGGCCGGCCAGAAAAUGCGU ....(.((..((((..((((.(.(((...((((.....)))).))).)))))..))))..)).)(((((.(((((((((((..((.((........)).)).)))))))))))..))))) ( -40.50) >DroYak_CAF1 3954 120 - 1 CAAACUCAUUUACUUGUAGCCAGGGCACUGCGGUACGUUUGCGGUCGUGUUAACGGUGUUGGCGGCGCAAUUCUGGCUGGUAGAAAUCAACACGUCCAAUUGGCCGGCCAGAAAAUGCGU ..................(((((.(((((((((.....)))))))(((....))))).))))).(((((.(((((((((((...(((...........))).)))))))))))..))))) ( -43.10) >consensus CAAACUCAUUUACUUGUAGCCAAGGCACUGCGGUACGUUUGCGGUCGUGUUAAUGGUGUUUGCGGCGCAAUUCUGGCUGGUAGAAAUCAACACGUCGAAUUGGCCGGCCAGAAAAUGCGC ..............(((.(((..(((((.((.(((....))).)).)))))...)))....)))(((((.(((((((((((.((...............)).)))))))))))..))))) (-35.08 = -34.34 + -0.75)



| Location | 6,589,336 – 6,589,456 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -26.72 |
| Energy contribution | -25.91 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6589336 120 + 22224390 AAACGUACCACAGUGCCUUGGCUACAAAUAAAUGAGUUUGAAUACGAUUUAAGUGGCAAUUUUCGGAAAAUAACUUUCUAGGCACAUGCACUCACCGGAAACGUGGAACGAAAAAGAAAG ...(((.((((.((((((..(((((...(((((..((......)).))))).))))).......(((((.....)))))))))))...........(....))))).))).......... ( -31.00) >DroSec_CAF1 4914 120 + 1 AAACGUACCGCAGUGCCUUAGCUACAAGUAAAUGAGUUUAAAUACGAUUUAAGUGACAAAUUUCGGAAAAUAACUUUCUGGGCACAUGCACUCACCGGAAACGUGGAACGAAAAAGAAAG ...(((.((((((((((.......((..(((((..((......)).)))))..))........((((((.....))))))))))).)))......((....)).)).))).......... ( -29.60) >DroEre_CAF1 4969 120 + 1 AAACGUACCGCAAUGCCCUGGCUACAAGUAAAUGAGUUUUGAAACGUUUUAAGUGGCAAUUUACGGAAAAUAACUCGCUGGGCACAUGCACUCACCGGAAACGUGGAACGAAAAAGAAAG ...(((.(((((.(((((.(((((....))...(((((......(((...((((....)))))))......)))))))))))))..)))......((....)).)).))).......... ( -29.00) >DroYak_CAF1 4034 120 + 1 AAACGUACCGCAGUGCCCUGGCUACAAGUAAAUGAGUUUGGAAACGUUUAAAGUGGCAAUUUACGGAAAAUAACUCGCUGGGCACAUGCACUCACCGGAAACGUGGAACGAAAAAGAAAG ...(((.(((((((((((..(((((...((((((..........))))))..)))))......(((........)))..)))))).)))......((....)).)).))).......... ( -34.00) >consensus AAACGUACCGCAGUGCCCUGGCUACAAGUAAAUGAGUUUGAAAACGAUUUAAGUGGCAAUUUACGGAAAAUAACUCGCUGGGCACAUGCACUCACCGGAAACGUGGAACGAAAAAGAAAG ...(((.((((.((((((.(((...........(((((......(((...............)))......))))))))))))))...........(....))))).))).......... (-26.72 = -25.91 + -0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:36 2006