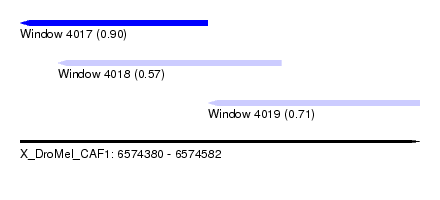
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,574,380 – 6,574,582 |
| Length | 202 |
| Max. P | 0.904197 |

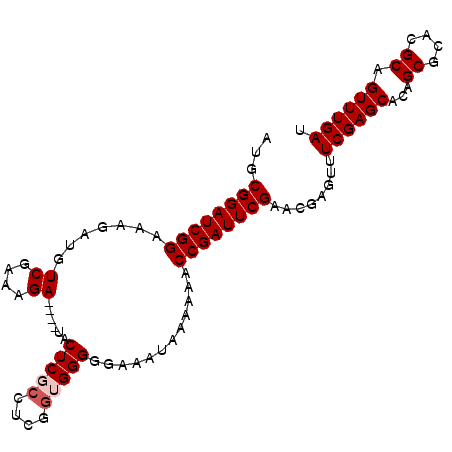
| Location | 6,574,380 – 6,574,475 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -23.54 |
| Energy contribution | -24.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6574380 95 - 22224390 AUGCGGAUCGGAAAGAUGUCGAAAGA----UACUCGACUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGCACAGCGCACGCAGUUUGAU .((((..((((((...((((....))----)).((((.(((((..............))))).)))).....))))))((.....)).))))....... ( -27.34) >DroSec_CAF1 19177 95 - 1 AUGCGGAUCGGAAAGAUGUCGAAAGA----UACUCGCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGCACAGCGCACGCAGUUUGAU ...((((((((.....((((....))----))(((((....)))))............))))))))........((((((...((....)).)))))). ( -29.60) >DroSim_CAF1 30956 95 - 1 AUGCGGAUCGGAAAGAUGUCGAAAGA----UACUCGCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGCACAGCGCACGCAGUUUGAU ...((((((((.....((((....))----))(((((....)))))............))))))))........((((((...((....)).)))))). ( -29.60) >DroEre_CAF1 18841 95 - 1 AUACGGAUCGGAAAGAUGUCGAAAGA----UACUCUCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGCACAGCGCACGCAGUUUGAU ...((((((((...((((((....))----)).))(((((....))))).........))))))))........((((((...((....)).)))))). ( -29.80) >DroYak_CAF1 28942 99 - 1 AUGCGGAUCGGAAAGAUGUCGAAAGAUAUAUACUCUCCUUGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGCACAGCGCACGCAGUUUGAU ...((((((((....(((((....)))))....((((((....)))))).........))))))))........((((((...((....)).)))))). ( -31.40) >consensus AUGCGGAUCGGAAAGAUGUCGAAAGA____UACUCGCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGCACAGCGCACGCAGUUUGAU ...((((((((.......((....))......(((((....)))))............))))))))........((((((...((....)).)))))). (-23.54 = -24.14 + 0.60)


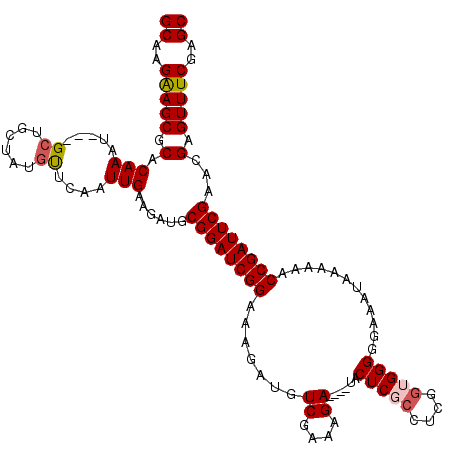
| Location | 6,574,399 – 6,574,512 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6574399 113 - 22224390 GCAAGAAGCGCACAAAU---GCUGCUACGUUCAAUUGAAGAUGCGGAUCGGAAAGAUGUCGAAAGA----UACUCGACUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGC ((..(((((((((....---).)))..(((((....(......)(((((((.....((((....))----))(((.((...)).)))...........)))))))))))).)))))..)) ( -31.50) >DroSec_CAF1 19196 113 - 1 GCAAGGAGCGCACAAAU---GCUGCUAUGCUCAAUUGAAGAUGCGGAUCGGAAAGAUGUCGAAAGA----UACUCGCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGC (((((.((((......)---))).)).)))....((((((...((((((((.....((((....))----))(((((....)))))............))))))))......)))))).. ( -36.00) >DroSim_CAF1 30975 113 - 1 GCAAGGAGCGCACAAAU---GCUGCUAUGCUCAAUUGAAGAUGCGGAUCGGAAAGAUGUCGAAAGA----UACUCGCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGC (((((.((((......)---))).)).)))....((((((...((((((((.....((((....))----))(((((....)))))............))))))))......)))))).. ( -36.00) >DroEre_CAF1 18860 113 - 1 GCAAGAAGCGCACAAAU---GCUGGCCUGUUCAAUUGAAGAUACGGAUCGGAAAGAUGUCGAAAGA----UACUCUCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGC ((..((((((((....)---)).....(((((....(......)(((((((...((((((....))----)).))(((((....))))).........)))))))))))).)))))..)) ( -32.60) >DroYak_CAF1 28961 120 - 1 GCAAGAAGCGCACAAAUGAUGCUGGUCUCUUUAAUUGAAGAUGCGGAUCGGAAAGAUGUCGAAAGAUAUAUACUCUCCUUGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGC ((..(((((((((....).)))..((.(((((....))))).))(((((((....(((((....)))))....((((((....)))))).........)))))))......)))))..)) ( -36.20) >consensus GCAAGAAGCGCACAAAU___GCUGCUAUGUUCAAUUGAAGAUGCGGAUCGGAAAGAUGUCGAAAGA____UACUCGCCUCGGUGGGGGAAAUAAAAAACCGAUUCGAACGAGUUUCGAGC ((..(((((.(.(((.....((......))....)))......((((((((.......((....))......(((((....)))))............))))))))...).)))))..)) (-24.46 = -24.78 + 0.32)


| Location | 6,574,475 – 6,574,582 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.70 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -20.68 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6574475 107 - 22224390 UUAUUUCCCACAU------UCGCACAUCAUCUUCAAUUGAAAGUGGUCAAUUAAUUGAAAUGUUUGUGCAGCGAUC----GCAAGAAGCGCACAAAU---GCUGCUACGUUCAAUUGAAG .............------...........(((((((((((.((((((((....))))...((((((((.((..((----....)).))))))))))---....)))).))))))))))) ( -32.50) >DroSec_CAF1 19272 107 - 1 UCGUUGACCACAU------UCGCACAUCGUCUUCAAUUGAAAGUGGUCAAUUAAUUGAAAUGUUUGUGCAGCGAUC----GCAAGGAGCGCACAAAU---GCUGCUAUGCUCAAUUGAAG ...((((((((.(------(((...............)))).))))))))((((((((...((((((((.((..((----....)).))))))))))---((......)))))))))).. ( -34.36) >DroSim_CAF1 31051 107 - 1 UCGUUGACCACAU------UCGCACAUCGUCUUCAAUUGAAAGUGGUCAAUUAAUUGAAAUGUUUGUGCAGCGAUC----GCAAGGAGCGCACAAAU---GCUGCUAUGCUCAAUUGAAG ...((((((((.(------(((...............)))).))))))))((((((((...((((((((.((..((----....)).))))))))))---((......)))))))))).. ( -34.36) >DroEre_CAF1 18936 106 - 1 UCGUUUCGCACAUCG-------CACAUUGUCUUCAAUUGAAAGUGGUCAAUUAAUUGAAAUGUUUGUGAAGCGAUC----GCAAGAAGCGCACAAAU---GCUGGCCUGUUCAAUUGAAG .......((.....)-------).......(((((((((((...((((((((......)))(((((((..((..((----....)).)).)))))))---..)))))..))))))))))) ( -30.70) >DroYak_CAF1 29041 120 - 1 UCGUUUCGCACAUCGCGCAUCGCACGUCGUCUUCAAUUGAAAGUGGUCAAUUAAUUGAAAUGUUUGUGCAGCGAUCAGCAGCAAGAAGCGCACAAAUGAUGCUGGUCUCUUUAAUUGAAG .((...(((.....)))...))........(((((((((((.(.(..((....(((.....((((((((.((.....)).((.....)))))))))))))..))..).)))))))))))) ( -29.40) >consensus UCGUUUCCCACAU______UCGCACAUCGUCUUCAAUUGAAAGUGGUCAAUUAAUUGAAAUGUUUGUGCAGCGAUC____GCAAGAAGCGCACAAAU___GCUGCUAUGUUCAAUUGAAG ..............................(((((((((((.((((((((....))))...((((((((.((...............)))))))))).......)))).))))))))))) (-20.68 = -21.60 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:17 2006