
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,571,077 – 6,571,217 |
| Length | 140 |
| Max. P | 0.999879 |

| Location | 6,571,077 – 6,571,177 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -27.80 |
| Energy contribution | -27.48 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.38 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6571077 100 + 22224390 GUAGCGCCAUAAAAACGCACUCGCUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAA ...(((.........))).......(((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).))))))). ( -27.50) >DroSec_CAF1 15988 100 + 1 GUAGCGCCAUAAAAACGCACUCGCUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAGUUAAAGCAGCGCAACUGGAA ...(((.........))).......(((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).))))))). ( -27.50) >DroSim_CAF1 26533 100 + 1 GUAGCGCCAUAAAAACGCACUCGCUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAA ...(((.........))).......(((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).))))))). ( -27.50) >DroEre_CAF1 15680 100 + 1 UUAGCGCCAUAAAAACGCACUUGUUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAA ...(((.........)))......((((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).)))))))) ( -27.60) >DroYak_CAF1 25549 101 + 1 UUAGCGCCAUAAAAACGCACUCGUUUCCAGUUUUGCUGUUUAAACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAGGCGCAAUUAAAGCAGCGCAACUGGAA ...(((.........)))......((((((((.((((((((.......((......)).(((((((((......)).))))))))))))))).)))))))) ( -27.40) >consensus GUAGCGCCAUAAAAACGCACUCGCUUCCAGUUUUGCUGUUUA_ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAA ...(((.........)))......((((((((.((((((((.......((......)).((((((((((....))).))))))))))))))).)))))))) (-27.80 = -27.48 + -0.32)



| Location | 6,571,077 – 6,571,177 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6571077 100 - 22224390 UUCCAGUUGCGCUGCUUUAAUUGCGCUUGAAAAAGACAAUCAUUCUAAUGCGACUUGU-UAAACAGCAAAACUGGAAGCGAGUGCGUUUUUAUGGCGCUAC ((((((((..((((.((((((..(((..(((...........)))....)))....))-))))))))..))))))))......(((((.....)))))... ( -28.80) >DroSec_CAF1 15988 100 - 1 UUCCAGUUGCGCUGCUUUAACUGCGCUUGAAAAAGACAAUCAUUCUAAUGCGACUUGU-UAAACAGCAAAACUGGAAGCGAGUGCGUUUUUAUGGCGCUAC ((((((((..((((.((((((..(((..(((...........)))....)))....))-))))))))..))))))))......(((((.....)))))... ( -30.30) >DroSim_CAF1 26533 100 - 1 UUCCAGUUGCGCUGCUUUAAUUGCGCUUGAAAAAGACAAUCAUUCUAAUGCGACUUGU-UAAACAGCAAAACUGGAAGCGAGUGCGUUUUUAUGGCGCUAC ((((((((..((((.((((((..(((..(((...........)))....)))....))-))))))))..))))))))......(((((.....)))))... ( -28.80) >DroEre_CAF1 15680 100 - 1 UUCCAGUUGCGCUGCUUUAAUUGCGCUUGAAAAAGACAAUCAUUCUAAUGCGACUUGU-UAAACAGCAAAACUGGAAACAAGUGCGUUUUUAUGGCGCUAA ((((((((..((((.((((((..(((..(((...........)))....)))....))-))))))))..))))))))......(((((.....)))))... ( -28.80) >DroYak_CAF1 25549 101 - 1 UUCCAGUUGCGCUGCUUUAAUUGCGCCUGAAAAAGACAAUCAUUCUAAUGCGACUUGUUUAAACAGCAAAACUGGAAACGAGUGCGUUUUUAUGGCGCUAA ...(((.....)))........((((((((((((((.......)))..(((.((((((((...(((.....))).))))))))))))))))).)))))... ( -26.70) >consensus UUCCAGUUGCGCUGCUUUAAUUGCGCUUGAAAAAGACAAUCAUUCUAAUGCGACUUGU_UAAACAGCAAAACUGGAAGCGAGUGCGUUUUUAUGGCGCUAC ((((((((..((((.((((((..(((..(((...........)))....)))....)).))))))))..))))))))......(((((.....)))))... (-25.32 = -25.16 + -0.16)


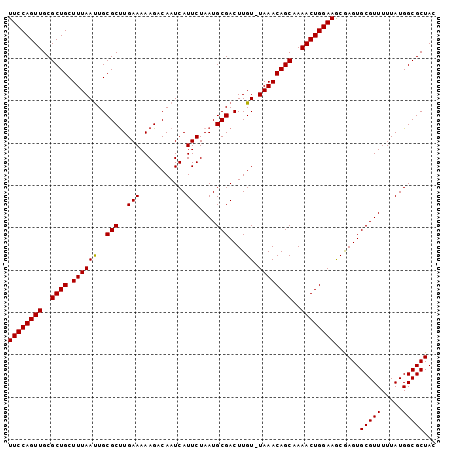
| Location | 6,571,098 – 6,571,217 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -32.42 |
| Energy contribution | -32.34 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6571098 119 + 22224390 CGCUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAAAAACUGCGUGUGAAUGGAAUACCCCGGACUUGAAAUGGUC (((((((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).)))))))).....)))......(((......)))((((......)))) ( -30.90) >DroSec_CAF1 16009 119 + 1 CGCUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAGUUAAAGCAGCGCAACUGGAAAAACUGCGUGUGAAUGGAAUACCCCGGACUUGACGUGGUC ...((((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).))))))))..((..(((..(..(((......)))..)...)))..)). ( -34.60) >DroSim_CAF1 26554 119 + 1 CGCUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAAAAACUGCGUGUGAAUGGAAUACCCCGGACUUGACGUGGUC ...((((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).))))))))..((..(((..(..(((......)))..)...)))..)). ( -34.60) >DroEre_CAF1 15701 119 + 1 UGUUUCCAGUUUUGCUGUUUA-ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAAAAACUGCGUGUGAAUGGAAUACCCCGGACUUGAUGUGGUC ..(((((((((.((((((((.-.....((......)).((((((((((....))).))))))))))))))).))))))))).((..(((..(..(((......)))..)...)))..)). ( -33.80) >DroYak_CAF1 25570 120 + 1 CGUUUCCAGUUUUGCUGUUUAAACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAGGCGCAAUUAAAGCAGCGCAACUGGAAAAACUGCGUGUGAAUGGAAUACCCCGGACUUGAUGUGGUC ..(((((((((.((((((((.......((......)).(((((((((......)).))))))))))))))).))))))))).((..(((..(..(((......)))..)...)))..)). ( -33.60) >consensus CGCUUCCAGUUUUGCUGUUUA_ACAAGUCGCAUUAGAAUGAUUGUCUUUUUCAAGCGCAAUUAAAGCAGCGCAACUGGAAAAACUGCGUGUGAAUGGAAUACCCCGGACUUGACGUGGUC ...((((((((.((((((((.......((......)).((((((((((....))).))))))))))))))).))))))))..(((((((..(..(((......)))..)...))))))). (-32.42 = -32.34 + -0.08)


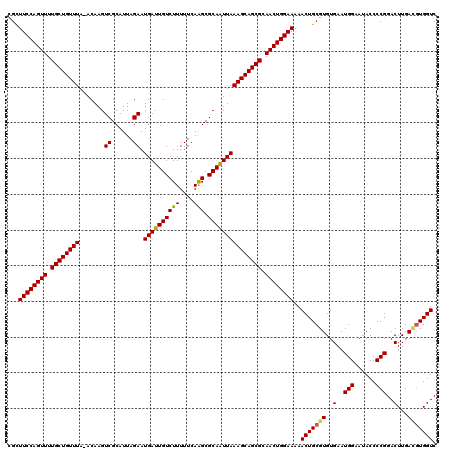
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:11 2006