
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,569,622 – 6,569,858 |
| Length | 236 |
| Max. P | 0.999262 |

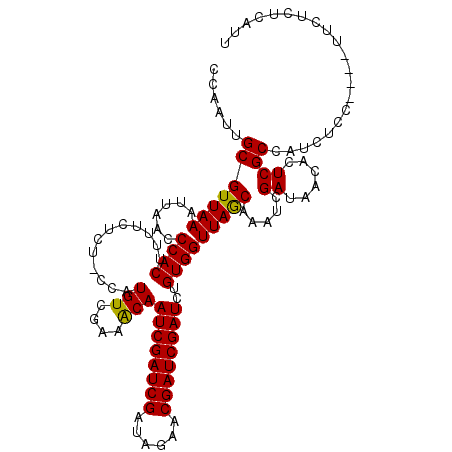
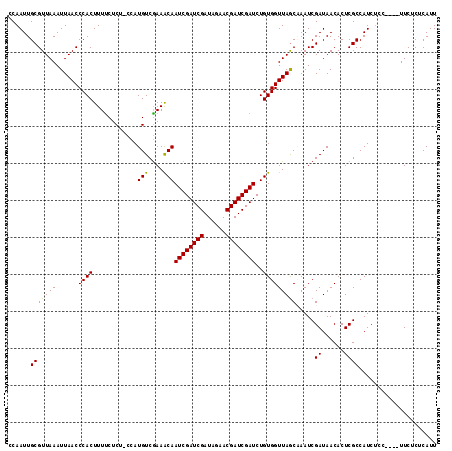
| Location | 6,569,622 – 6,569,738 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6569622 116 - 22224390 CCAAUUGCGUUAAAUUAACCCACUUUUCUCCCCCAUGUCGAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUAGCAAAUCGAUAACACUCGCCAUCUCCUUCCUUCUCUCAUU ....((((((((...))))((((............(((....)))((((((((......))))))))..))))...))))..(((......)))...................... ( -18.20) >DroSec_CAF1 14551 111 - 1 CCAAUUGCGUUAAAUUAACCCACUUUUCUCU-CCAUGUCGAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUAGCCAAUCGAUAACACUCGCCAUCUCC----UUCUCUCAUU ......((((((...))))............-...((((((.(((((((((((......)))))))).)))((....))..))))))......)).......----.......... ( -20.50) >DroSim_CAF1 25106 111 - 1 CCAAUUGCGUUAAAUUAACCCACUUUUCUCU-CCAUGUCAAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUAGCCAAUCGAUAACACUCGCCAUCUCC----UUCUCUCAUU .(.((((.(((((......((((........-...(((....)))((((((((......))))))))..))))))))))))).)..................----.......... ( -17.80) >DroEre_CAF1 14246 110 - 1 CCAAUUGCGUUAAAUUAACCCACUUUCCUC--CCAUGUCGAAGCAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUAACAAAUCGAUAACACUCGCCAUCCC----UUUCUCUCAUU ......((((((...))))...........--...((((((.(((((((((((......)))))))).)))(.....)...))))))......))......----........... ( -17.70) >consensus CCAAUUGCGUUAAAUUAACCCACUUUUCUCU_CCAUGUCGAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUAGCAAAUCGAUAACACUCGCCAUCUCC____UUCUCUCAUU ......(((((((......((((............(((....)))((((((((......))))))))..))))))))).....((......))))..................... (-16.80 = -16.43 + -0.37)

| Location | 6,569,659 – 6,569,778 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -27.72 |
| Energy contribution | -27.40 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6569659 119 + 22224390 UGC-UAACCACAGAUCGAUCGUUCUAUCGAUCGAUUGUUUCGACAUGGGGGAGAAAAGUGGGUUAAUUUAACGCAAUUGGGCGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGC (((-...((((.(((((((((......))))))))).((((..(.....)..)))).))))((((...))))))).....(((((....(((((((.((......)).)))))))))))) ( -37.30) >DroSec_CAF1 14584 118 + 1 GGC-UAACCACAGAUCGAUCGUUCUAUCGAUCGAUUGUUUCGACAUGG-AGAGAAAAGUGGGUUAAUUUAACGCAAUUGGGAGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGC (((-.....((((((((.((((((..((..(((((((((((..((...-.........((.((((...)))).))..)).)))))))))))...))..)))))).))))))))..))).. ( -35.84) >DroSim_CAF1 25139 118 + 1 GGC-UAACCACAGAUCGAUCGUUCUAUCGAUCGAUUGUUUUGACAUGG-AGAGAAAAGUGGGUUAAUUUAACGCAAUUGGGAGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGC (((-.....((((((((.((((((..((..(((((((((((..((...-.........((.((((...)))).))..)).)))))))))))...))..)))))).))))))))..))).. ( -33.94) >DroEre_CAF1 14279 117 + 1 UGU-UAACCACAGAUCGAUCGUUCUAUCGAUCGAUUGCUUCGACAUGG--GAGGAAAGUGGGUUAAUUUAACGCAAUUGGGCGGCAAUUGAGCAGAAUGAAUGAACGAUCUAUUCAUCGC ...-...((((.(((((((((......))))))))).((((......)--)))....)))).((((((...(((......)))..))))))((.((.((((((.......)))))))))) ( -31.00) >DroYak_CAF1 24175 117 + 1 UGUUUAAUCAGAGAUCGAUCGUUGUAUCGAUCGAUU-UUUCGACAUGG--GAGGAAAGUGGGUUAAUUUAACGCAAUUGGUCGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGC .........((((((((((((......)))))))))-)))((((....--.......((.(..(((((......)))))..).))....(((((((.((......)).))))))))))). ( -34.20) >consensus UGC_UAACCACAGAUCGAUCGUUCUAUCGAUCGAUUGUUUCGACAUGG__GAGAAAAGUGGGUUAAUUUAACGCAAUUGGGCGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGC .......((((.(((((((((......))))))))).((((.........))))...))))...........(((((((.....)))))(((((((.((......)).)))))))...)) (-27.72 = -27.40 + -0.32)

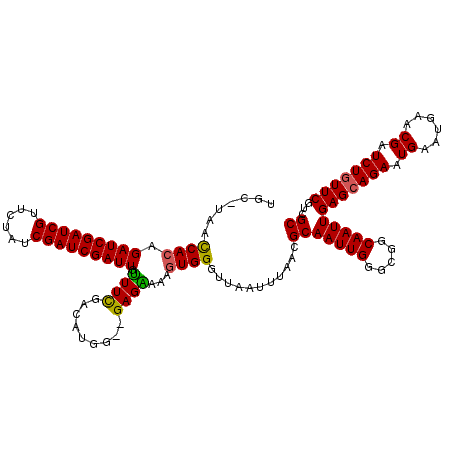
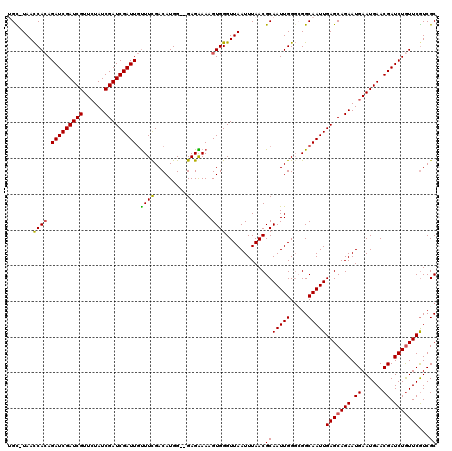
| Location | 6,569,659 – 6,569,778 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6569659 119 - 22224390 GCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGCCCAAUUGCGUUAAAUUAACCCACUUUUCUCCCCCAUGUCGAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUA-GCA (((((..((((((((.((.((((((..((.((((((.....)))))).))..)))....................((((((.........))))))))).)).)))))))).))).-)). ( -29.40) >DroSec_CAF1 14584 118 - 1 GCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCUCCCAAUUGCGUUAAAUUAACCCACUUUUCUCU-CCAUGUCGAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUA-GCC (((((..((((((((.((.((((((..((.((((((.....)))))).))..)))................-...((((((.........))))))))).)).)))))))).))).-)). ( -28.50) >DroSim_CAF1 25139 118 - 1 GCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCUCCCAAUUGCGUUAAAUUAACCCACUUUUCUCU-CCAUGUCAAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUA-GCC (((((((((.....))))....(((..((.((((((.....)))))).))..)))................-....)))...(((((((((((......)))))))).))).....-)). ( -25.50) >DroEre_CAF1 14279 117 - 1 GCGAUGAAUAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGCCCAAUUGCGUUAAAUUAACCCACUUUCCUC--CCAUGUCGAAGCAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUA-ACA ..(((((((.((....)))))))))..((.((((((.....)))))).))....((((((....(((..(--....)..)))(((((((((((......)))))))).))))))))-).. ( -25.80) >DroYak_CAF1 24175 117 - 1 GCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGACCAAUUGCGUUAAAUUAACCCACUUUCCUC--CCAUGUCGAAA-AAUCGAUCGAUACAACGAUCGAUCUCUGAUUAAACA .((((((((.....))))....(((..((.((((((.....)))))).))..)))...............--....))))...-.((((((((......))))))))............. ( -23.70) >consensus GCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGCCCAAUUGCGUUAAAUUAACCCACUUUUCUC__CCAUGUCGAAACAAUCGAUCGAUAGAACGAUCGAUCUGUGGUUA_GCA .((((((((.....))))....(((..((.((((((.....)))))).))..))).....................))))..(((((((((((......)))))))).)))......... (-23.26 = -23.18 + -0.08)

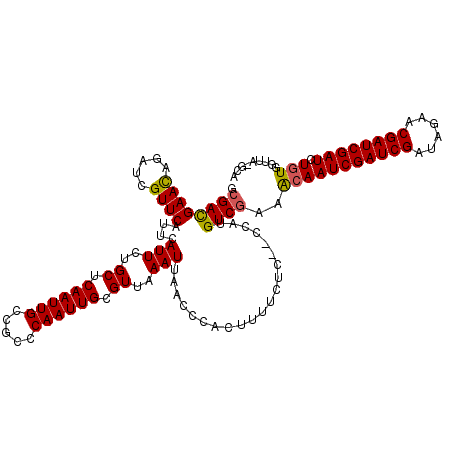
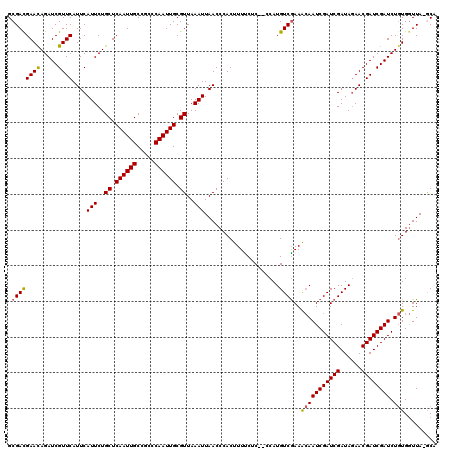
| Location | 6,569,738 – 6,569,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6569738 120 + 22224390 GCGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGCGCCCAAGCGACAUCGUAAUUGAUUAAUAAAAAAUUAUGCCCGAAAUAGUCGCGGGCACUGAAAUUAACCGUAAGUGCCGA (((((....(((((((.((......)).)))))))(((((......)))))..(((((((...........))))))).........))))).((((((.............)))))).. ( -38.52) >DroSec_CAF1 14662 120 + 1 GAGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGCGCACAAGCGACAUCGUAAUUGAUUAAUAAAAAAUUAUGCCCGAAAUAGUCGCGGGCACUGAAAUUAACCGUAAGUGCCGA ..((((...(((((((.((......)).)))))))(((((......))))).((((((((...........))))))(((((.........)))))...)).............)))).. ( -34.60) >DroSim_CAF1 25217 120 + 1 GAGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGCGCCCAGGCGACAUUGUAAUUGAUUAAUAAAAAAUUAUGCCCGAAAUAGUCGCGGGCACUGAAAUUAACCGUAAGUGCCGA ..((((...(((((((.((......)).)))))))(((((......))))).........................((((((.........)))))).................)))).. ( -33.70) >DroEre_CAF1 14356 118 + 1 GCGGCAAUUGAGCAGAAUGAAUGAACGAUCUAUUCAUCGCGCCCAAGCGACAUCGUAAUUGAUUAAUAAAAAAUUAUGCCCGAAAUAGUC--GGGCACUGAAAUUAACCGUAAGUGCCGA .(((((.((((.((((((.(((..(((((.......((((......)))).))))).))).)))............(((((((.....))--))))))))...)))).(....)))))). ( -32.20) >DroYak_CAF1 24252 120 + 1 UCGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGCGCCCAAGCGACAACGUAAUUGAUUAAUAAAAAAUUAUGCCCGAAAUAGUCGCGGGCACUGAAAUUAACCGUAAGUGCCGA ((((((...(((((((.((......)).)))))))(((((......))))).((((((((...........)))))((((((.........))))))...........)))...)))))) ( -37.40) >consensus GCGGCAAUUGAGCAGAAUGAAUGAACGAUCUGUUCGUCGCGCCCAAGCGACAUCGUAAUUGAUUAAUAAAAAAUUAUGCCCGAAAUAGUCGCGGGCACUGAAAUUAACCGUAAGUGCCGA ..((((...(((((((.((......)).)))))))(((((......))))).........................)))).............((((((.............)))))).. (-31.30 = -31.70 + 0.40)

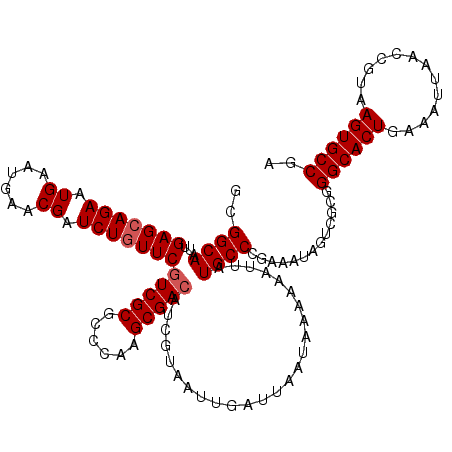
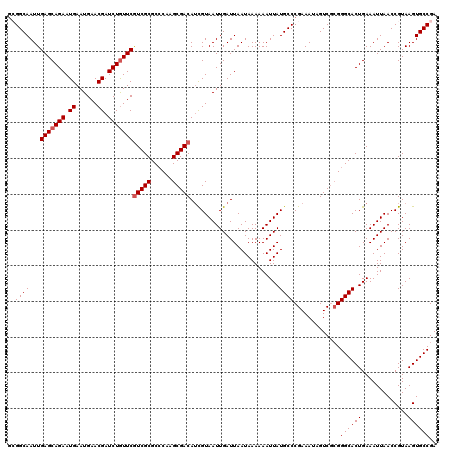
| Location | 6,569,738 – 6,569,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -30.92 |
| Energy contribution | -30.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6569738 120 - 22224390 UCGGCACUUACGGUUAAUUUCAGUGCCCGCGACUAUUUCGGGCAUAAUUUUUUAUUAAUCAAUUACGAUGUCGCUUGGGCGCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGC .(((((.....(((((((....(((((((.........)))))))........))))))).........(((((......)))))((.((((..(......)..)))).))...))))). ( -33.40) >DroSec_CAF1 14662 120 - 1 UCGGCACUUACGGUUAAUUUCAGUGCCCGCGACUAUUUCGGGCAUAAUUUUUUAUUAAUCAAUUACGAUGUCGCUUGUGCGCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCUC ..((((.....(((((((....(((((((.........)))))))........))))))).........(((((......)))))((.((((..(......)..)))).))...)))).. ( -32.00) >DroSim_CAF1 25217 120 - 1 UCGGCACUUACGGUUAAUUUCAGUGCCCGCGACUAUUUCGGGCAUAAUUUUUUAUUAAUCAAUUACAAUGUCGCCUGGGCGCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCUC ..((((.....(((((((....(((((((.........)))))))........))))))).........(((((......)))))((.((((..(......)..)))).))...)))).. ( -31.50) >DroEre_CAF1 14356 118 - 1 UCGGCACUUACGGUUAAUUUCAGUGCCC--GACUAUUUCGGGCAUAAUUUUUUAUUAAUCAAUUACGAUGUCGCUUGGGCGCGAUGAAUAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGC .(((((.....(((((((....((((((--((.....))))))))........)))))))..............(((((((.(((((((.((....))))))))).))))))).))))). ( -38.90) >DroYak_CAF1 24252 120 - 1 UCGGCACUUACGGUUAAUUUCAGUGCCCGCGACUAUUUCGGGCAUAAUUUUUUAUUAAUCAAUUACGUUGUCGCUUGGGCGCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGA ((((((.....(((((((....(((((((.........)))))))........)))))))......((((.(((....)))))))((.((((..(......)..)))).))...)))))) ( -35.20) >consensus UCGGCACUUACGGUUAAUUUCAGUGCCCGCGACUAUUUCGGGCAUAAUUUUUUAUUAAUCAAUUACGAUGUCGCUUGGGCGCGACGAACAGAUCGUUCAUUCAUUCUGCUCAAUUGCCGC ..((((.....(((((((....(((((((.........)))))))........))))))).........(((((......)))))((.((((..(......)..)))).))...)))).. (-30.92 = -30.80 + -0.12)

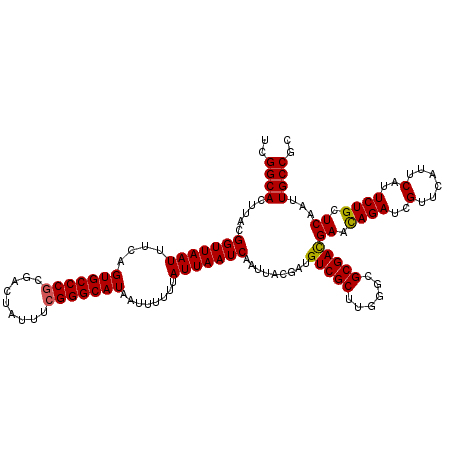
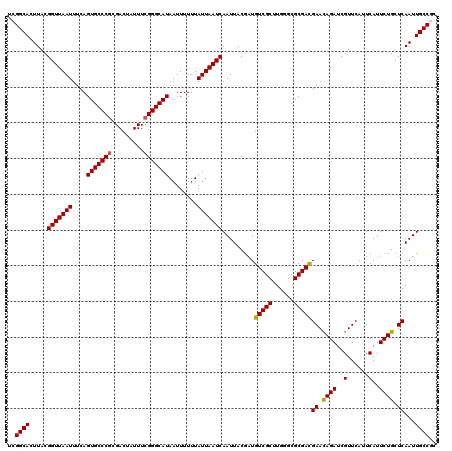
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:09 2006