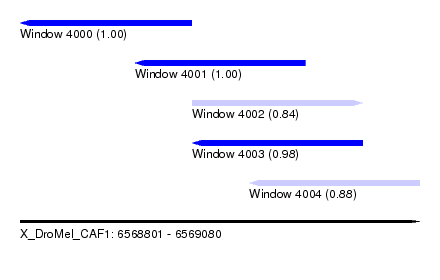
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,568,801 – 6,569,080 |
| Length | 279 |
| Max. P | 0.999951 |

| Location | 6,568,801 – 6,568,921 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -47.34 |
| Consensus MFE | -47.18 |
| Energy contribution | -47.30 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.79 |
| SVM RNA-class probability | 0.999951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568801 120 - 22224390 UAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAGGAAAUGGAUGUCAUAUCCAUGUCCUUCAUUCUGCCAUCGGCGGAGUGUGACCUGGAUUUGAAUACGGAGACCAAGGGCUG .....(((((((((((....)).))))))))).((((..((...((....(((.(((((.(((...(((((((((...))))))))).))).))))).)))...))....))..)))).. ( -49.20) >DroSec_CAF1 13721 120 - 1 UAUACUGCUGGCCAUCUGCGGAUUGGUCAGUACGUCCGAGGAAAUGGAUGUCAUAUCCAUGUCCUUCAUUCUGCCAUCGGCGGAGUGUGACCUGGAUUUGAAUACGGAGACCAAGGGCUG .....(((((((((((....)).))))))))).((((..((...((....(((.(((((.(((...(((((((((...))))))))).))).))))).)))...))....))..)))).. ( -47.20) >DroSim_CAF1 24277 120 - 1 UAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAGGAAAUGGAUGUCAUAUCCAUGUCCUUCAUUCUGCCAUCGGCGGAGUGUGACCUGGAUUUGAAUACGGAGACCAAGGGCUG .....(((((((((((....)).))))))))).((((..((...((....(((.(((((.(((...(((((((((...))))))))).))).))))).)))...))....))..)))).. ( -49.20) >DroEre_CAF1 13448 120 - 1 UAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAGGAAAUGGAUGUCAUCUCCAUGUCCUUCAUUCUGCCCUCGGCUGAGUGUGACCUGGAUUUGAACACGGAGACCAAGGGCUG .....(((((((((((....)).))))))))).((((..((...((....(((..((((.(((...(((((.(((...))).))))).))).))))..)))...))....))..)))).. ( -44.30) >DroYak_CAF1 23334 120 - 1 CAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAGGAAAUGGAUGUCAUCUCCAUGUCCUUCAUUCUGCCAUCGGCGGAAUGUGAUCUGGAUUUGAAUACGGAGACCAAGGGCUG .....(((((((((((....)).))))))))).((((..((...((....(((..((((.(((...(((((((((...))))))))).))).))))..)))...))....))..)))).. ( -46.80) >consensus UAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAGGAAAUGGAUGUCAUAUCCAUGUCCUUCAUUCUGCCAUCGGCGGAGUGUGACCUGGAUUUGAAUACGGAGACCAAGGGCUG .....(((((((((((....)).))))))))).((((..((...((....(((.(((((.(((...(((((((((...))))))))).))).))))).)))...))....))..)))).. (-47.18 = -47.30 + 0.12)



| Location | 6,568,881 – 6,569,000 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -45.04 |
| Consensus MFE | -44.40 |
| Energy contribution | -44.08 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568881 119 - 22224390 CCU-UGCCGGAACCCACUGCCCCAAUAGUCGCCAAUUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUAUAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAG .((-((..(((((....((((...(((((((((((((.(((........)))))))).)))))))))))).))))).......(((((((((((((....)).))))))))).)).)))) ( -46.10) >DroSec_CAF1 13801 120 - 1 CCUUUGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUAUAUACUGCUGGCCAUCUGCGGAUUGGUCAGUACGUCCGAG ........(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).......(((((((((((((....)).))))))))).))..... ( -42.90) >DroSim_CAF1 24357 119 - 1 CCU-UGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUAUAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAG .((-((..(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).......(((((((((((((....)).))))))))).)).)))) ( -46.10) >DroEre_CAF1 13528 119 - 1 CCU-UGCCGGAACCCACUGCCCCGAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUAUAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAG .((-((..(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).......(((((((((((((....)).))))))))).)).)))) ( -46.10) >DroYak_CAF1 23414 119 - 1 CCU-UGCCGGAACCCACUGCCCCGAUAGUCGCCAACUUUGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUACAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAG .((-((..(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).......(((((((((((((....)).))))))))).)).)))) ( -44.00) >consensus CCU_UGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUAUAUACUGCUGGCCAUCUGCGGAUUGGUCAGCACGUCCGAG ........(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).......(((((((((((((....)).))))))))).))..... (-44.40 = -44.08 + -0.32)



| Location | 6,568,921 – 6,569,040 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -33.06 |
| Energy contribution | -33.74 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568921 119 + 22224390 UAAUGGAACUUGCCAUAGCCGCACAAUUCGAUGGCCCUCUCGAAAUUGGCGACUAUUGGGGCAGUGGGUUCCGGCA-AGGGUACGGACUUCUUGCGUCCCUUACCACCCUUGCCCCCCUU ....((((((((((((((.(((.((((((((........))).)))))))).))))...))))...))))))((((-(((((..((((.......))))......)))))))))...... ( -46.10) >DroSec_CAF1 13841 120 + 1 UAAUGGAACUUGCCAUAGCCGCACAAUUCGAUGGCCCUCUCGAAGUUGGCGACUAUUGGGGCAGUGGGUUCCGGCAAAGGGUACGGACUUCUUGCGUCCCUUAGCACCCAUGCCCCCCUU ....((........((((.(((.((((((((........))).)))))))).)))).((((((.(((((....((.(((((.(((.(.....).)))))))).))))))))))))))).. ( -44.70) >DroSim_CAF1 24397 119 + 1 UAAUGGAACUUGCCAUAGCCGCACAAUUCGAUGGCCCUCUCGAAGUUGGCGACUAUUGGGGCAGUGGGUUCCGGCA-AGGGUACGGACUUCUUGCGUCCCUUACCACCCUUGCCCCCCUU ....((((((((((((((.(((.((((((((........))).)))))))).))))...))))...))))))((((-(((((..((((.......))))......)))))))))...... ( -46.10) >DroEre_CAF1 13568 110 + 1 UAAUGGAACUUGCCAUAGCCGCACAAUUCGAUGGCCCUCUCGAAGUUGGCGACUAUCGGGGCAGUGGGUUCCGGCA-AGGGUACGGACUUCUUGCGUCCCUUG---------CCCCCCUU ....((((((((((((((.(((.((((((((........))).)))))))).))))...))))...))))))((((-((((.(((.(.....).)))))))))---------))...... ( -44.30) >DroYak_CAF1 23454 107 + 1 UAAUGGAACUUGCCAUAGCCGCACAAUUCGAUGGCCCUCUCAAAGUUGGCGACUAUCGGGGCAGUGGGUUCCGGCA-AGGGUAUGGACUUCUUGCGUCCCUUA---------CCCC---U ....((..((((((..((((.(((..(((((((((((..........)).).))))))))...)))))))..))))-))((((.((((.......))))..))---------))))---. ( -38.90) >consensus UAAUGGAACUUGCCAUAGCCGCACAAUUCGAUGGCCCUCUCGAAGUUGGCGACUAUUGGGGCAGUGGGUUCCGGCA_AGGGUACGGACUUCUUGCGUCCCUUA_CACCC_UGCCCCCCUU ....((((((((((((((.(((.((((((((........))).)))))))).))))...))))...))))))((((.((((.(((.(.....).))))))).........))))...... (-33.06 = -33.74 + 0.68)


| Location | 6,568,921 – 6,569,040 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -49.42 |
| Consensus MFE | -37.20 |
| Energy contribution | -39.04 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568921 119 - 22224390 AAGGGGGGCAAGGGUGGUAAGGGACGCAAGAAGUCCGUACCCU-UGCCGGAACCCACUGCCCCAAUAGUCGCCAAUUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUA ..((((((((.(((((((((((((((.........))).))))-))))...))))..)))))).(((((((((((((.(((........)))))))).))))))))........)).... ( -53.30) >DroSec_CAF1 13841 120 - 1 AAGGGGGGCAUGGGUGCUAAGGGACGCAAGAAGUCCGUACCCUUUGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUA ......((((.((((((....((((.......))))))))))..))))(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).... ( -51.00) >DroSim_CAF1 24397 119 - 1 AAGGGGGGCAAGGGUGGUAAGGGACGCAAGAAGUCCGUACCCU-UGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUA ..((((((((.(((((((((((((((.........))).))))-))))...))))..)))))).(((((((((((.(((((........)))))))).))))))))........)).... ( -53.30) >DroEre_CAF1 13568 110 - 1 AAGGGGGG---------CAAGGGACGCAAGAAGUCCGUACCCU-UGCCGGAACCCACUGCCCCGAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUA ......((---------(((((((((.........))).))))-))))(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).... ( -47.20) >DroYak_CAF1 23454 107 - 1 A---GGGG---------UAAGGGACGCAAGAAGUCCAUACCCU-UGCCGGAACCCACUGCCCCGAUAGUCGCCAACUUUGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUA (---((((---------((..((((.......)))).))))))-)...(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).... ( -42.30) >consensus AAGGGGGGCA_GGGUG_UAAGGGACGCAAGAAGUCCGUACCCU_UGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCGAGAGGGCCAUCGAAUUGUGCGGCUAUGGCAAGUUCCAUUA ......((((.(((.....(.((((.......)))).)..))).))))(((((....((((...(((((((((((.(((((........)))))))).)))))))))))).))))).... (-37.20 = -39.04 + 1.84)


| Location | 6,568,961 – 6,569,080 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -55.76 |
| Consensus MFE | -44.18 |
| Energy contribution | -44.92 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568961 119 - 22224390 CGCCCCCGAUGGGGGUCACGGGGGUCACGGGCCGCCCGGCAAGGGGGGCAAGGGUGGUAAGGGACGCAAGAAGUCCGUACCCU-UGCCGGAACCCACUGCCCCAAUAGUCGCCAAUUUCG .((((((...))))))...((((((.....))).)))(((....((((((.(((((((((((((((.........))).))))-))))...))))..)))))).......)))....... ( -57.00) >DroSec_CAF1 13881 120 - 1 CGCCCCCGAUGGGGGUCACGGGGGUCACGGGACGCCCGGCAAGGGGGGCAUGGGUGCUAAGGGACGCAAGAAGUCCGUACCCUUUGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCG .((((((...))))))....(((((...(((....(((((((((((((((....))))...((((.......))))...)))))))))))..)))...)))))................. ( -55.90) >DroSim_CAF1 24437 119 - 1 CGCCCCCGAUGGGGGUCACGGGGGUCACGGGCCGCCCGGCAAGGGGGGCAAGGGUGGUAAGGGACGCAAGAAGUCCGUACCCU-UGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCG .((((((...))))))...((((((.....))).)))(((....((((((.(((((((((((((((.........))).))))-))))...))))..)))))).......)))....... ( -57.00) >DroEre_CAF1 13608 110 - 1 CGCCCCCGAUGGGGGUCACGGGGGUCAUGGGCCGCCCGGCAAGGGGGG---------CAAGGGACGCAAGAAGUCCGUACCCU-UGCCGGAACCCACUGCCCCGAUAGUCGCCAACUUCG .((((((...))))))....(((((..((((....((((((((((..(---------(...((((.......)))))).))))-))))))..))))..)))))((.(((.....))))). ( -54.30) >DroYak_CAF1 23494 107 - 1 CGCCCCCGAUGGGGGUCACGGGGGUCAUGGGCCACCCGGUA---GGGG---------UAAGGGACGCAAGAAGUCCAUACCCU-UGCCGGAACCCACUGCCCCGAUAGUCGCCAACUUUG .((((((...))))))....(((((..((((....((((((---((((---------((..((((.......)))).))))))-))))))..))))..)))))................. ( -54.60) >consensus CGCCCCCGAUGGGGGUCACGGGGGUCACGGGCCGCCCGGCAAGGGGGGCA_GGGUG_UAAGGGACGCAAGAAGUCCGUACCCU_UGCCGGAACCCACUGCCCCAAUAGUCGCCAACUUCG .((((((...))))))....(((((...(((....((((((.((((.((....))......((((.......))))...)))).))))))..)))...)))))................. (-44.18 = -44.92 + 0.74)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:04 2006